Geospatial information
Several different sensors include geospatial information in the dataset metadata describing the location of the sensor at the time of capture.
Coordinate reference systems The Scanalyzer system itself does not have a reliable GPS unit on the sensor box. There are 3 different coordinate systems that occur in the data:
Most common is EPSG:4326 (WGS84) USDA coordinates
Tractor planting & sensor data is in UTM Zone 12
Sensor position information is captured relative to the southeast corner of the Scanalyzer system in meters
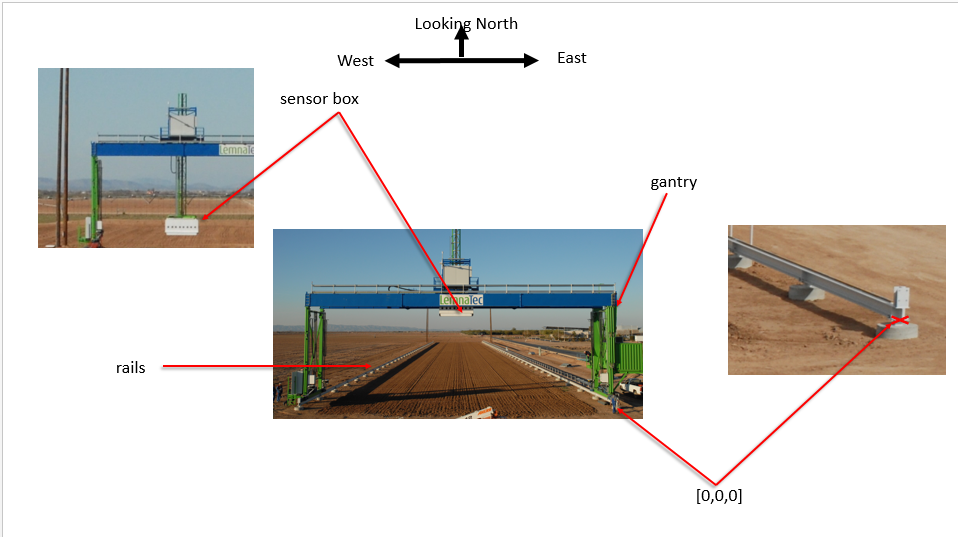
EPSG:4326 coordinates for the four corners of the Scanalyzer system (bound by the rails above) are as follows:
NW: 33° 04.592' N, -111° 58.505' W
NE: 33° 04.591' N, -111° 58.487' W
SW: 33° 04.474' N, -111° 58.505' W
SE: 33° 04.470' N, -111° 58.485' W
In the trait database, this site is named the "MAC Field Scanner Field" and its bounding polygon is "POLYGON ((-111.9747967 33.0764953 358.682, -111.9747966 33.0745228 358.675, -111.9750963 33.074485715 358.62, -111.9750964 33.0764584 358.638, -111.9747967 33.0764953 358.682))"
Scanalyzer coordinates Finally, the Scanalyzer coordinate system is right-handed - the origin is in the SE corner, X increases going from south to north, and Y increases from east to the west.
In offset meter measurements from the southeast corner of the Scanalyzer system, the extent of possible motion for the sensor box is defined as:
NW: (207.3, 22.135, 5.5)
SE: (3.8, 0, 0)
Scanalyzer -> EPSG:4326 1. Calculate the UTM position of known SE corner point 2. Calculate the UTM position of the target point, using SE point as reference 3. Get EPSG:4326 position based on UTM
MAC coordinates Tractor planting data and tractor sensor data will use UTM Zone 12.
Scanalyzer -> MAC Given a Scanalyzer(x,y), the MAC(x,y) in UTM zone 12 is calculated using the linear transformation formula:
Assume Gx = -Gx', where Gx' is the Scanalyzer X coordinate.
MAC -> Scanalyzer
MAC -> EPSG:4326 USDA We do a linear shifting to convert MAC coordinates in to EPSG:4326 USDA
Sensors with geospatial metadata
stereoTop
flirIr
co2
cropCircle
PRI
scanner3dTop
NDVI
PS2
SWIR
VNIR
Available data All listed sensors
stereoTop
cropCircle
co2Sensor
flirIrCamera
ndviSensor
priSensor
SWIR
field scanner plots
There are 864 (54*16) plots in total and the plot layout is described in the plot plan table.
dimension
value
# rows
32
# rows / plot
2
# plots (2 rows ea)
864
# ranges
54
# columns
16
row width (m)
0.762
plot length (m)
4
row length (m)
3.5
alley length (m)
0.5
The boundary of each plot changes slightly each planting season. The scanalyzer coordinates of each row and each range of the two planting seasons is available in the field book. The scanalyzer coordinates of each plot are transformed into the (EPSG:4326) USDA coordinates using the equations above. After that, a polygon of each plot can be generated using ST_GeomFromText funtion and inserted into the BETYdb through SQL statements.
An Rcode is available for generating SQL statements based on the scanalyzer coordinates of each plot, which takes range.csv and row.csv as standard inputs.
The range.csv should be in the following format:
range
x_south
x_north
1
...
...
2
...
...
3
...
...
...
...
...
And the row.csv should look like:
row
y_west
y_east
1
...
...
2
...
...
3
...
...
...
...
...
The output will be something look like:
Last updated