Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
This user manual is divided into the following sections:
Data Products: A summary of the available data products and the processes used to create them
Data Access: Instructions for how to access the data products using Clowder, Globus, BETYdb, and CoGe
Description of the scientific objectives and experimental design
Data use policy: Information about data use and attribution
User Tutorials: In-depth examples of how to access and use the TERRA-REF data
Raw output from sensors deployed on Lemnatec field and greenhouse systems, UAVs and tractors
Manually-collected fieldbooks and associated protocols
Derived data, including phenomics data, from computational approaches
Genomic pipeline data
The TERRA-REF reference dataset may be of interest to a variety of research communities including:
Computer vision\/remote sensing\/image analysis (raw sensor data and metadata)
Physiologists (plants and how they are growing)
Robotics (gantry\/location\/orientation)
Breeders (derived traits)
Genomics\/bioinformaticians (genomics data)
The following table lists available TERRA-REF data products. The table will be updated as new datasets are released. Links are provided to pages with detailed information about each data product including sensor descriptions, algorithm (extractor) information, protocols, and data access instructions.
Infrared heat imaging data is collected collected using the FLIR SC615 thermal sensor. These data are provided as geotiff image raster files as well as plot level means.
Algorithms are in the repository; see the readme for details.
Sensor information:
ua-mac/Level_1/ir_geotiff
To be created
Plot level summaries are named in the trait database. In the future this name will be used for the Level 1 data as well. This name from the Climate Forecast (CF) conventions, and is used instead of 'canopy_temperature' for two reasons: First, because we do not (currently) filter soil in this pipeline. Second, because the CF definition of surface_temperature distinguishes the surface from the medium: "The surface temperature is the temperature at the interface, not the bulk temperature of the medium above or below."
Thermal imaging data is available via Clowder and Globus:
/ua-mac/raw_data/flirIrCamera
Data are unavailable for Season 4 (summer 2017 sorghum) and season 5 (winter 2017-2018 wheat).
Hyperspectral imaging data is collected using the Headwall VNIR and SWIR sensors. In the Nov 2017 Beta Release only VNIR data is provided because we do not have the measurements of downwelling spectral radiation required by the pipeline.
Please see the for more information about how the data are generated and known issues.
See
Hyperspectral data is available via Clowder, , the , and our :
Clowder:
SWIR Collection: Level 1 data not available
Globus and Workbench:
VNIR: /sites/ua-mac/Level_1/vnir_netcdf
SWIR: Level 1 data not available
Sensor information:
Level 2 data are spectral indices computed at the same resolution as Level 1. These can be found in the same Level 1 directories as their parents, but the files are appended *_ind.nc.
To get a list of hyperspectral indices currently generated:
Raw data is available in the filesystem, accessible via Workbench and Globus in the following directories:
VNIR: /sites/ua-mac/raw_data/VNIR
SWIR: /sites/ua-mac/raw_data/SWIR
These files are uncalibrated; see the hyperspectral pipeline repository for information on how these can be processed.
Fluorescence intensity data is collected using the PSII camera.
Fluorescence intensity data is available via Clowder and Globus:
Clowder:
Globus path: /sites/ua-mac/raw_data/ps2top
Sensor information:
For details about using this data via Clowder or Globus, please see section.
Description: Raw image output is converted to a raster format (netCDF\/GeoTIFF)
Output: /sites/ua_mac/Level_1/ps2top
There are 102 bin files. The first (index 0) is an image taken right before the LED are switched on (dark reference). Frame 1 to 100 are the 100 images taken, with the LEDs on. In binary file 102 (index 101) is a list with the timestamps of each frame of the 100 frames.
Right now the LED on timespan is 1s thus the first 50 frames are taken with LEDs on the latter 50 frames with LED off..
3D point cloud data is collected using the Fraunhofer 3D laserscanner. .
Data is available via Clowder and Globus.
Clowder:
Globus path: /sites/ua_mac/raw_data/scanner3DTop
Sensor information:
For details about using this data via Clowder or Globus, please see section.
Raw sensor output (PLY) is converted to LAS format using the ply2las extractor
Description: PLY data is converted to LAS using the 3D point cloud extractor
Output:
Clowder: LAS file is added to the dataset
Globus: /sites/ua_mac/Level_1/scanner3DTop
Genomic data includes whole-genome resequencing data from the HudsonAlpha Institute for Biotechnology, Alabama for 384 samples for accessions from the sorghum (BAP) and genotyping-by-sequencing (GBS) data from Kansas State University for 768 samples from a population of sorghum recombinant inbred lines (RIL).
These data are available to Beta Users and require permission to access. The form to sign up for our beta user program is at . Once you have signed up for our beta user program you can access genomics data in one of the following locations:
Download via .
The , which provides container-based computing environments including Jupyter, Rstudio, and Python IDE.
The for download or use within the CyVerse computing environment.
The computing environment.
See before continuing.
The data is structured on both the TERRA-REF strorage (accessible via Globus and Workbench) and CyVerse Data Store infrastructures as follows:
Data derived from analysis of the raw resequencing data at the Danforth Center (version1) are available as gzipped, genotyped variant call format (gVCF) files and the final combined hapmap file.
Combined genotype calls are available in VCF format.
For details about using this data via Clowder or Globus, please see section.
Work to recover these data is ongoing; see
Problem description
For details about using this data via Clowder or Globus, please see section.
Raw data are in bzip2 FASTQ format, one per read pair (*_R1.fastq.bz2 and *_R2.fastq.bz2). 384 samples are available. For a list of the lines sequenced, see the .
Raw data are in gzip FASTQ format. 768 samples are available. For a list of lines sequenced, see the .
Data product
Description
3D point cloud data (LAS) of the field constructed from the Fraunhofer 3D scanner output (PLY).
Fluorescence intensity imaging is collected using the PSII LemnaTec camera. Raw camera output is converted to (netCDF/GeoTIFF)
Hyperspectral imaging data from the SWIR and VNIR Headwall Inspector sensors are converted to netCDF output using the hyperspectral extractor.
Infrared heat imaging data is collected using FLIR sensor. Raw output is converted to GeoTIFF using the FLIR extractor.
Multispectral data is collected using the PRI and NDVI Skye sensors. Raw output is converted to timeseries data using the multispectral extractor.
Stereo imaging data is collected using the Prosilica cameras. Full-color images are reconstructed in GeoTIFF format using the de-mosaic extractor. A full-field mosaic is generated using the full-field mosaic extractor.
Spectral reflectance data
Spectral reflectance is measured using a Crop Circle active crop canopy sensor
Environment conditions are collected through the CO2 sensor and Thies Clima. Raw output is converted to netCFG using the environmental-logger extractor.
postGIS/netCDF
Phenotype data is derived from sensor output using the PlantCV extractor and imported into BETYdb.
FASTQ and VCF files available via Globus
UAV and Phenotractor
Plot level data available in BETYdb
Interested researchers can access BETYdb directly from GIS software such as ESRI ArcMap and QGIS. In some cases direct access can simplify the use of spatial data in BETYdb data, but this convenience must be weighed against a more complex setup, limits of GIS software compatibility, and additional complexity of extracting data from a PostGIS SQL database.
Accessing the production BETYdb used by the TERRA REF program requires creating a secure shell tunnel (SSH) to a remote server. After creating the tunnel, the database is accessed as if it were available on the local machine. A step-by-step process is given below.
ArcMap 10.3 or later (Requires Windows operating system)
Instructions for using QGIS and other GIS software are provided below
PuTTY: ssh client for Windows that can be downloaded here: PuTTY
Request access to the BETYdb server by following the link. This will take you to the NCSA identity service. If you do not have an NCSA account, you will be asked to create one. This account and password will be used to login to the database server. Access will generally be granted within 24-hours.
Use PuTTY or your preferred SSH client and your NCSA account. First open the terminal and then login to bety6.ncsa.illinois.edu using ssh from the command line:
After confirming access to bety6 logout by typing exit.
The following command will create an SSH tunnel from your computer to the BETYdb server:
Note if have a postgres running on your desktop computer (using the default port 5432), you will need to stop it first.
The above will bind the local port 5432 (first parameter) to port 5432 (second parameter), the default Postgres listening port, on the remote server. All traffic bound for port 5432 on your local machine will be automatically forwarded to the remote server. As a result, programs such as ArcGIS running on your computer will connect to the remote BETYdb as if it were on your computer.
Note you will need to create the SSH connection with the tunnel every time you wish to access BETYdb from your local machine.
To keep the tunnel open, use
note for PuTTY Users: you can configure Putty to remember these settings. In the navigation tree on the left-hand side, click Connection > SSH > Tunnels. Enter '5432' under Source port and 'localhost:5432' in the Destination field. Then click session and save this configuration for future use.
The next section of the guide will discuss accessing BETYdb using ArcMap, querying plots and joining these to the traits and experiments tables. The instructions for setting up a SSH tunnel will also work psql, pgAdmin3, QGIS, and other clients. Instructions for connecting via QGIS and ArcGIS Pro are provided below.
BETYdb is configured with PostGIS geometry support. This allows ArcGIS Desktop clients to access geometry layers stored within BETYdb.
Warning: ArcGIS releases prior to 10.3 required you to place the PostgreSQL libpq files in the ArcGIS client's bin directory. This is no longer required for the ArcGIS Desktop clients but some ESRI tools may still require the library be installed.*
Click on the ArcCatalog icon (on right edge of ArcMap window) to open the ArcCatalog Tree
In the tree, click on 'Database Connections' and then "Add Database Connnections". A Database Connection dialog window will open.
Within the dialog box:
Click OK
The connection will be saved as "Connection to localhost.sde", right
click and rename to it to "TERRA REF BETYdb trait database" to allow easy reuse.
Click on the Add Layer icon (black cross over yellow diamand) button to open the Add Data dialog window.
Under 'Look in' on the second line choose 'Database Connections'.
Select the "TERRA REF BETYdb trait database" that created above
Select the bety.public.sites table and click 'Add'.
This 'sites' table is the only table in the database with a geospatial 'geometry' data type.
Any of the other tables can also be added, as described below.
The New Query Layer dialog will be displayed asking for the Unique Identifier Field for the layer. For the bety.public.sites table, the unique identifier is the "sitename" field.
Click Finish.
Warning: ArcMap does not support the big integer format used by BETYdb as primary keys and those fields will not be visible or available for selection. In most cases you should be able to use other fields as unique identifiers.*
BETYdb contains one geometry table called betydb.public.sites containing the boundaries for each plot. Because the plot boundaries can change each season, and even within season, different plot definitions may be used (e.g. to subset plots or exclude boundary rows), there is significant overlap that can cause confusion when displayed. In general, you will want to use the query layer to limit plots to a single season and a single definition.
Right click the bety.public.sites layer and choose properties.
Choose the Definition Query tab
Add the line sitename LIKE 'MAC Field Scanner Season 1%' or sitename LIKE 'MAC Field Scanner Season 2%' to limit the layer to Season 1 or Season 2 respectively.
Click 'OK'
For more advanced selection of sites by experiment or season, you can join the experiments and experiments_sites tables. This is beyond the scope of the present tutorial.
Additional tables can be added and joined to the sites table. Tables can be added just like any other layer. In this case, we'll add bety.public.traits_and_yields_view and join it to the bety.public.sites layer.
To create a join with other tables, start by adding the desired table.
Follow instructions above to add the bety.public.traits_and_yields_view
On this table the unique identifier is a group of columns, so select sitename, cultivar, scientificname, trait, date, entity, and method as the unique identifiers.
Right click on the bety.public.sites layer.
Under 'Joins and Relates' select 'Join'.
Choose sitename (from bety.public.sites) in part 1
Choose bety.public.traits_and_yields_view in part 2
Choose sitename in part 3
Click OK
The final section describes how to create a thematic view of the bety.public.sites layer based on the mean attribute where the trait is NDVI from the bety.public.traits_and_yields_view. Remove any previous joins from bety.public.sites (right click bety.public.sites --> joins and relates --> remove join) prior to performing this procedure because we will be selecting the NDVI data by creating a query layer from bety.public.traits_and_yields_view prior to the join.
Right click bety.public_traits_and_yields_view table and select properties
Click on the Definition Query tab
Add the line "trait = 'NDVI'" to the Definition Query box
Click OK
Follow the steps defined in Joining Additional BETYdb Tables
Right click on the bety.sites layer and choose properties
Choose the Symbology tab
Under the Show section, choose Quantities --> Graduated Colors
Under the Fields Value selection choose mean
Click OK
Below connection instructions assume an SSH tunnel exists.
This assumes you have followed instructions for ArcMAP to create a database connection file.
Open ArcCatalog
Under database connections, you will find the connection made above, called 'TERRA REF BETYdb.sde'
right click this and select 'properties'
copy the file path (it should look like C:\Users\<USER NAME>\AppData\Roaming\ESRI\Desktop10.4\ArcCatalog\TERRA REF BETYdb.sde
Open ArcGIS Pro
Under the Insert tab, select connections --> 'add database'
paste the path to 'TERRA REF BETYdb.sde' in the directory navigation bar
select 'TERRA REF BETYdb.sde'
Open QGIS
In left 'browser panel', right-click the PostGIS icon
select 'New Connection'
Enter connection properties
Name: TERRA REF BETYdb trait database
Service: blank
Host: localhost
Port: 5432
Database: bety
SSL mode: disable
Username: viewer
Password: DelchevskoOro
Options: select 'Also list tables with no geometry'
This does not require GIS software other than the PostGIS traits database. While connecting directly to the database within GIS software is handy, it is also straightforward to export Shapefiles.
After you have connected via ssh to the PostGIS server, the pgsql2shp function is available and can be used to dump out all of the plot and site definitions (names and geometries) thus:
Real-time sensor data transfer by file number and size can be viewed here.
See Data Products for more information about individual data products and How to Access Data for instructions to access the data products.
BETYdb is used to manage and distribute agricultural and ecological data. It contains phenotype and agronomic data including plot locations and other geolocations of interest (e.g. fields, rows, plants).
To request access to BETYdb, register on the BETYdb web site. You will be notified once you have been granted access.
The primary BETYdb Data Access Guide is largely relevant here, noting the following usages:
Genotypes are stored in the cultivars table
Plots are stored in the sites table. Plots are nested hierarchically based on geolocation.
Most tables in BETYdb have search boxes. We describe below how to use the Advanced Search box to query data from these tables and download the results as a CSV file.
The Advanced Search box is the easiest way to download summary datasets designed to have enough information (location, time, species, citations) to be useful for a wide range of use cases.
(For more information about querying data from specific tables, see the BETYdb Data Access Guide.)
On the Welcome page of BETYdb there is a search option for trait and yield data (Figure 1). This tool allows users to search the entire collection of trait and yield data for specific sites, citations, species, and traits.
The results page provides a map interface and the option to download a file containing search results. The downloaded file is in CSV format. This file provides meta-data and provenance information, including the SQL query used to extract the data, the date and time the query was made, the citation source of each result row, and a citation for BETYdb itself.
Using the search box to search trait and yield data is very simple: Type the site (city or site name), species (scientific or common name), cultivar, citation (author and/or year), or trait (variable name or description) into the search box and the results will show contents of BETYdb that match the search. The number of records per page can be changed to accord with the viewer's preference and the search results can be downloaded in the Excel-compatible CSV format.
The search map may be used in conjunction with search terms to restrict search results to a particular geographical area—or even a specific site—by clicking on a map. Clicking on a particular site will restrict results to that site. Clicking in the vicinity of a group of sites but not on a particular site will restrict the search to the region around the point clicked. Alternatively, if a search using search terms is done without clicking on the map, all sites associated with the returned results are highlighted on the map. Then, to zero in on results for a particular geographic area, click on or near highlighted locations on the map.
Produced with Gitbook version
Environment conditions data is collected using the Vaisala CO2, Thies Clima weather sensors as well as lightning, irrigation, and weather data collected at the Maricopa site.
Data formats follow the Climate and Forecast (CF) conventions for variable names and units. Environmental data are stored in the Geostreams database.
WeatherStation coordinates are 33.074457 N, 111.975163 W
EnvironmentLogger is on top of the gantry system and is moveable.
Irrigation is managed at the field level. There are four regions that can be irrigated at different rates.
Level 1 meteorological data is aggregated to from 1 Hz raw data to 5 minute averages or sums.
On Globus or Workbench you can find these data provided in both hourly and daily files. These files contain data at the original temporal resolution of 1/s. In addition, they contain the high resolution spectral radiometer data.
sites/ua-mac/Level_1/envlog_netcdf
hourly files: YYYY-MM-DD_HH-MM-SS_environmentallogger.nc
daily files: envlog_netcdf_L1_ua-mac_YYYY-MM-DD.nc
Data can be accessed using the geostreams API or the PEcAn meteorological workflow. These are illustrated in the sensor data tutorials.
Here is the json representation of a single five-minute observation:
Data can be accessed using the geostreams API or the PEcAn meteorological workflow.
These are illustrated in the sensor data tutorials.
Here is the json representation of a single five-minute observation from Geostreams:
Data is available via Globus or Workbench:
/ua-mac/raw_data/co2sensor
/ua-mac/raw_data/EnvironmentLogger
/ua-mac/raw_data/irrigation
/ua-mac/raw_data/lightning
/ua-mac/raw_data/weather
Description: EnvironmentalLogger raw files are converted to netCDF.
Known issue: the irrigation data stream does not currently handle variable irrigation rates within the field. Specifically, we have not yet accounted for the Summer 2017 drought experiments. See terraref/reference-data#196 for more information.
When the full field is irrigated (as is typical), the irrigated area is 5466.1 m2 (=215.2 m x 25.4 m)
In 2017:
Full field irrigated area from the start of the season to August 1 (103 dap) is 5466.1 m2 (=215.2 m x 25.4 m).
Well-watered treatment zones from August 1 to 15 (103 to 116 dap): 2513.5 m2 (=215.2 m x 11.68 m) in total, combined areas of non-contiguous blocks
Well-watered treatment zones from August 15 - 30 (116 to 131 dap): 3169.9 m2 (=215.2 m x 14.73 m), again in total as the combined areas of non-contiguous blocks
Several different sensors include geospatial information in the dataset metadata describing the location of the sensor at the time of capture.
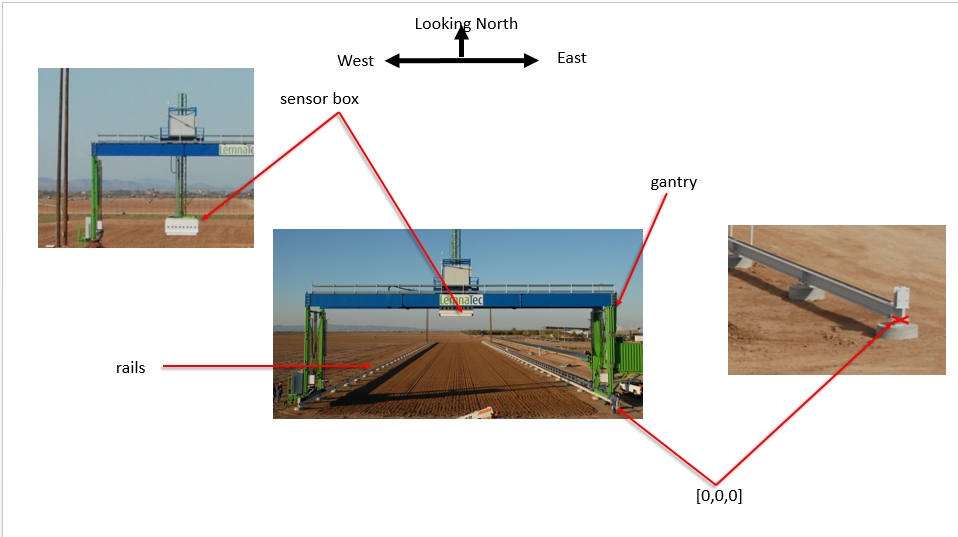
Coordinate reference systems The Scanalyzer system itself does not have a reliable GPS unit on the sensor box. There are 3 different coordinate systems that occur in the data:
Most common is EPSG:4326 (WGS84) USDA coordinates
Tractor planting & sensor data is in UTM Zone 12
Sensor position information is captured relative to the southeast corner of the Scanalyzer system in meters
EPSG:4326 coordinates for the four corners of the Scanalyzer system (bound by the rails above) are as follows:
NW: 33° 04.592' N, -111° 58.505' W
NE: 33° 04.591' N, -111° 58.487' W
SW: 33° 04.474' N, -111° 58.505' W
SE: 33° 04.470' N, -111° 58.485' W
In the trait database, this site is named the "MAC Field Scanner Field" and its bounding polygon is "POLYGON ((-111.9747967 33.0764953 358.682, -111.9747966 33.0745228 358.675, -111.9750963 33.074485715 358.62, -111.9750964 33.0764584 358.638, -111.9747967 33.0764953 358.682))"
Scanalyzer coordinates Finally, the Scanalyzer coordinate system is right-handed - the origin is in the SE corner, X increases going from south to north, and Y increases from east to the west.
In offset meter measurements from the southeast corner of the Scanalyzer system, the extent of possible motion for the sensor box is defined as:
NW: (207.3, 22.135, 5.5)
SE: (3.8, 0, 0)
Scanalyzer -> EPSG:4326 1. Calculate the UTM position of known SE corner point 2. Calculate the UTM position of the target point, using SE point as reference 3. Get EPSG:4326 position based on UTM
MAC coordinates Tractor planting data and tractor sensor data will use UTM Zone 12.
Scanalyzer -> MAC Given a Scanalyzer(x,y), the MAC(x,y) in UTM zone 12 is calculated using the linear transformation formula:
Assume Gx = -Gx', where Gx' is the Scanalyzer X coordinate.
MAC -> Scanalyzer
MAC -> EPSG:4326 USDA We do a linear shifting to convert MAC coordinates in to EPSG:4326 USDA
Sensors with geospatial metadata
stereoTop
flirIr
co2
cropCircle
PRI
scanner3dTop
NDVI
PS2
SWIR
VNIR
Available data All listed sensors
stereoTop
cropCircle
co2Sensor
flirIrCamera
ndviSensor
priSensor
SWIR
field scanner plots
There are 864 (54*16) plots in total and the plot layout is described in the plot plan table.
The boundary of each plot changes slightly each planting season. The scanalyzer coordinates of each row and each range of the two planting seasons is available in the field book. The scanalyzer coordinates of each plot are transformed into the (EPSG:4326) USDA coordinates using the equations above. After that, a polygon of each plot can be generated using ST_GeomFromText funtion and inserted into the BETYdb through SQL statements.
An Rcode is available for generating SQL statements based on the scanalyzer coordinates of each plot, which takes range.csv and row.csv as standard inputs.
The range.csv should be in the following format:
And the row.csv should look like:
The output will be something look like:
Meteorological data will use Climate Forecasting 'standard names' and 'canonical units' conventions. CF is widely used in climate, meteorology, and earth sciences.
Here are some examples (note that we can change from canonical units to match the appropriate scale, e.g. "C" instead of "K"; time can use any base time and time step (e.g. hours since 2015-01-01 00:00:00 UTC, etc. But the time zone has to be UTC, where 12:00:00 is approx (+/- 15 min). solar noon at Greenwich.
standard_name is CF-convention standard names (except irrigation)
units can be converted by udunits, so these can vary (e.g. the time denominator may change with time frequency of inputs)
Before the Running
The pipepline is developed in Python, so a Python Interpreter is a must. Other than the basic Python standard librarys, the following third-party libraries are required:
netCDF4 for Python
numpy
Other than official CPython interpreter, Pypy is also welcomed, but please make sure that these third-party modules are correctly installed for the target interpreter. The pipeline can only works in Python 2.X versions (2.7 recommended) since numpy does not support Python 3.X versions.
Cloning from the Git:
The extractor for this pipeline is developed and maintained by Max in branch "EnvironmentalLogger-extractor" under the same repository.
Get the Environmental Logger Pipeline to Work
To trigger the pipeline, use the following command:
python ${environmental_logger_source_path}/environmental_logger_json2netcdf.py ${input_JSON_file} ${output_netCDF_file}
Where:
${environmental_logger_source_path} is where the three environmental_logger files are located
${input_JSON_file} is where the input JSON files are located
${output_netCDF_file} is where the users want the pipeline to export the product (netCDF file)
Please note that the parameter for the output file can be a path to either a directory or a file, and it is not necessarily to be existed. If the output is a path to a folder, the final product will be in this folder as a netCDF file that has the same name as the imported JSON file but with a different filename extension (.nc for standard netCDF file); if this path does not exist, environmental_logger pipeline will automatically make one.
The calculation in the Environmental Logger is mainly finished by the module environmental_logger_calculation.py under the support of numpy.
TERRA-REF data is available through four different approaches: Globus Connect, Clowder, BETYdb, and CoGe. Raw data is transfered to the primary compute pipeline using Globus Online. Data is ingested into Clowder to support exploratory analysis. The Clowder extractor system is used to transform the data and create derived data products, which are either available via Clowder or published to specialized services, such as BETYdb.
For more information, see the .
Clowder is the primary system used to organize, annotate, and process raw data generated by the phenotyping platforms as well as information about sensors.
Use Clowder to explore the raw TERRA-REF data, perform exploratory analysis, and develop custom extractors.
For more information, see .
Raw data is transferred to the primary TERRA-REF compute pipeline on the (ROGER) system using Globus Online. Data is available for Globus transfer via the . Direct access to ROGER is restricted.
Use Globus Online when you want to transfer data from the TERRA-REF system for local analysis.
For more information, see .
BETYdb contains the derived trait data with plot locations and other information associated with agronomic experimental design.
Use BETYdb to access about derived trait data.
CoGe contains genomic information and sequence data.
Field protocols
Calibration protocols
Clowder is an active data repository designed to enable collaboration around a set of shared datasets. TERRAREF uses Clowder to organize, annotate, and process data generated by phenotyping platforms. Datafiles are available via the Clowder or .
See the for more information about the software and its applications.
To create an account, sign up at the and wait for your account to be approved. Once access is granted, you can explore collections and datasets.
Data is organized into spaces, collections, and datasets, collections.
Spaces contain collections and datasets. TERRA-REF uses one space for each of the phenotyping platforms.
Collections consist of one or more datasets. TERRA-REF collections are organized by acquisition date and sensor. Users can also create their own collections.
Datasets consist of one or more files with associated metadata collected by one sensor at one time point. Users can annotate, download, and use these sensor datasets.
Clowder allows users to search metadata and filter datasets and files with particular attributes. Simply enter your search terms in the search box.
Clowder includes support for launching integrated analysis environments from your browser, including RStudio and Jupyter Notebooks.
After selecting a dataset, under the "Analysis Environment Instances", select the "Launch new instance with dataset" drop-down, select the desired tool, then the "Launch" button. Select the "Environment manager" link to view the list of active instances. Find your instance and select the title link. This will display the tool with the selected dataset mounted. If you have a running instance, you can also "Upload dataset to existing instance".
The Analysis Workbench allows you to launch private Jupyter Notebook and RStudio instances to explore and analyze TERRA-REF data products.
To create an account, sign up at the TERRA-REF site and wait for your account to be approved. Once access is granted, you can launch analysis environments.
Each user has a "home" directory mounted into the analysis tools under /home/userid. This is read-write scratch space.
Data access is provided via a read-only NFS mount to the TERRA-REF dataset on ROGER. The data is mounted to each container under /data/terraref and linked to the analysis environment working directory. For example, in Jupyter this is /home/jovyan/work/data.
is a National Science Foundation funded cyberinfrastructure that aims to democratize access to supercomputing capabilities.
TERRA-REF genomics data is accessible on the CyVerse Data Store and Discovery Environment. Accessing data through the CyVerse Discovery Environment requires signing up for a free CyVerse account. The Discovery Environment gives users access to software and computing resources, so this method has the advantage that TERRA-REF data can be utilized directly without the need to copy the data elsewhere. During the TERRA-REF , users will need to request access to the TERRA-REF CyVerse Community Data folder through the TERRA-REF . The TERRA-REF Community Data folder can be found at /iplant/home/shared/terraref.
The service provides high-performance, secure, file transfer and synchronization between endpoints. It also allows you to securely share your data with other Globus users.
To access data via Globus, you must first have a Globus account and endpoint.
Sign up for Globus at
Download and install Globus Connect or
To request access to the Terraref endpoint, send your Globus id (or University email) to David LeBauer (dlebauer@illinois.edu) with 'TERRAREF Globus Access Request' in the subject. You will be notified once you have been granted access.
To transfer data to your computer or server:
Log into Globus
Add an endpoint for the destination (e.g. your local computer)
Go to the 'transfer files' page:
Select source
Endpoint: Terraref
Path: Navigate to the subdirectory that you want.
Select (click) a folder
Select (highlight) files that you want to download at destination
Select the endpoint that you set up above of your local computer or server
Select the destination folder (e.g. /~/Downloads/)
Click 'go'
Files will be transfered to your computer
For more information, see .
For more information, see .
Field scanner operational log
Through it's extractor architecture, Clowder supports automated computational workflows. For more information about developing Clowder extractors, see the documentation
Globus
dimension
value
# rows
32
# rows / plot
2
# plots (2 rows ea)
864
# ranges
54
# columns
16
row width (m)
0.762
plot length (m)
4
row length (m)
3.5
alley length (m)
0.5
range
x_south
x_north
1
...
...
2
...
...
3
...
...
...
...
...
row
y_west
y_east
1
...
...
2
...
...
3
...
...
...
...
...
CF standard-name
units
time
days since 1700-01-01 00:00:00 UTC
air_temperature
K
air_pressure
Pa
mole_fraction_of_carbon_dioxide_in_air
mol/mol
moisture_content_of_soil_layer
kg m-2
soil_temperature
K
relative_humidity
%
specific_humidity
1
water_vapor_saturation_deficit
Pa
surface_downwelling_longwave_flux_in_air
W m-2
surface_downwelling_shortwave_flux_in_air
W m-2
surface_downwelling_photosynthetic_photon_flux_in_air
mol m-2 s-1
precipitation_flux
kg m-2 s-1
irrigation_flux
kg m-2 s-1
irrigation_transport
kg s-1
wind_speed
m/s
eastward_wind
m/s
northward_wind
m/s
CF standard-name
units
bety
isimip
cruncep
narr
ameriflux
air_temperature
K
airT
tasAdjust
tair
air
TA (C)
air_temperature_max
K
tasmaxAdjust
NA
tmax
air_temperature_min
K
tasminAdjust
NA
tmin
air_pressure
Pa
air_pressure
PRESS (KPa)
mole_fraction_of_carbon_dioxide_in_air
mol/mol
CO2
moisture_content_of_soil_layer
kg m-2
soil_temperature
K
soilT
TS1 (NOT DONE)
relative_humidity
%
relative_humidity
rhurs
NA
rhum
RH
specific_humidity
1
specific_humidity
NA
qair
shum
CALC(RH)
water_vapor_saturation_deficit
Pa
VPD
VPD (NOT DONE)
surface_downwelling_longwave_flux_in_air
W m-2
same
rldsAdjust
lwdown
dlwrf
Rgl
surface_downwelling_shortwave_flux_in_air
W m-2
solar_radiation
rsdsAdjust
swdown
dswrf
Rg
surface_downwelling_photosynthetic_photon_flux_in_air
mol m-2 s-1
PAR
PAR (NOT DONE)
precipitation_flux
kg m-2 s-1
cccc
prAdjust
rain
acpc
PREC (mm/s)
degrees
wind_direction
WD
wind_speed
m/s
Wspd
WS
eastward_wind
m/s
eastward_wind
CALC(WS+WD)
northward_wind
m/s
northward_wind
CALC(WS+WD)
We plan to make data from the Transportation Energy Resources from Renewable Agriculture Phenotyping Reference Platform (TERRA-REF) project available for use with attribution. Each type of data will include or point to the appropriate attribution policy.
We plan to release the data in stages or tiers. For pre-release access please complete the alpha tester application.
The first tier will be an internal release to the TERRA-REF team and the standards committee. This first tier release will be to initially quality check and calibrate the data and will take place as data sets are produced and compiled.
By November 2016, it is an objective of the TERRA-REF team to establish a data release pipeline, wherein the release of data to this first tier will be within 21 days from the date of collection.
Access to the data will be arranged for by the resource producer (i.e. limiting access to selected users).
The second tier will enable the release of the data generated solely by the TERRA-REF team to other TERRA teams as well as non-TERRA entities.
By November 2017, it is an objective of the TERRA-REF team to establish a data release pipeline, wherein the release of data to this second tier will be within 10 days from the data of collection.
It is noted that release of the data to the second tier may occur prior to publication and that access is granted with the understanding that the contributions and interests of the TERRA-REF team should be recognized and respected by the users of the data. The TERRA-REF team reserves the right to analyze and published its own data. Resource users should appropriately cite the source of the data and acknowledge the resource produces. The publication of the data, as suggested in the TERRA-REF Authorship Guidelines, should specify the collaborative nature of the project, and authorship is expected to include all those TERRA-REF team members contributing significantly to the work.
Access to the data will be determined by the resource producers and may be governed by separate license or other agreements. 1. iii)It is an objective of the TERRA-REF team to enable the release of the data to the public by November 2018 but no later than the date of close-out of the awarded funds.
Genomic data for the Sorghum bicolor Bioenergy Association Panel (BAP) from the TERRA-REF project is available pre-publication to maximize the community benefit of these resources. Use of the raw and processed data that is available should follow the principles of the Fort Lauderdale Agreement and the Department of Energy's Joint Genome Institute (JGI) early release policies.
By accessing these data, you agree not to publish any articles containing analyses of genes or genomic data on a whole genome or chromosome scale prior to publication by TERRA-REF and/or its collaborators of a comprehensive genome analysis ("Reserved Analyses"). "Reserved analyses" include the identification of complete (whole genome) sets of genomic features such as genes, gene families, regulatory elements, repeat structures, GC content, or any other genome feature, and whole-genome- or chromosome-scale comparisons with other species. The embargo on publication of Reserved Analyses by researchers outside of the TERRA-REF project is expected to extend until the publication of the results of the sequencing project is accepted. Scientific users are free to publish papers dealing with specific genes or small sets of genes using the sequence data. If these data are used for publication, the following acknowledgment should be included: 'These sequence data were produced by the US Department of Energy Transportation Energy Resources from Renewable Agriculture Phenotyping Reference Platform (TERRA-REF) Project'. These data may be freely downloaded and used by all who respect the restrictions in the previous paragraphs. The assembly and sequence data should not be redistributed or repackaged without permission from TERRA-REF. Any redistribution of the data during the embargo period should carry this notice: "The TERRA-REF project provides these data in good faith, but makes no warranty, expressed or implied, nor assumes any legal liability or responsibility for any purpose for which the data are used. Once the sequence is moved to unreserved status, the data will be freely available for any subsequent use."
We prefer that potential users of these sequence data contact the individuals listed under Contacts with their plans to ensure that proposed usage of sequence data are not considered Reserved Analyses.
For algorithms, we intend to release via MIT or MIT compatible license (e.g. BSD, UIUC/NCSA, Apache v2).
For other raw data, such as phenotypic data and associated metadata, we intend to release via Creative Commons with Attribution (CC by 4.0).
Todd Mockler, Project/Genomics Lead (email: tmockler AT danforthcenter DOT org)
David LeBauer, Computing Pipeline Lead (email: dlebauer AT illinois DOT edu)
Erica Fishel, Technology Transfer Lead (email: efischel AT danforthcenter DOT org)
Nadia Shakoor, Associate Project Director (email: nshakoor AT danforthcenter DOT org)
We will release the data in stages or tiers.
The first tier will be an internal release to the TERRA-REF team and the standards committee. This first tier release will be to initially quality check and calibrate the data and will take place as data sets are produced and compiled.
By November 2016, it is an objective of the TERRA-REF team to establish a data release pipeline, wherein the release of data to this first tier will be within 21 days from the date of collection.
Access to the data will be arranged for by the resource producer (i.e. limiting access to selected users).
The second tier will enable the release of the data generated solely by the TERRA-REF team to other TERRA teams as well as non-TERRA entities.
By November 2017, it is an objective of the TERRA-REF team to establish a data release pipeline, wherein the release of data to this second tier will be within 10 days from the data of collection.
It is noted that release of the data to the second tier may occur prior to publication and that access is granted with the understanding that the contributions and interests of the TERRA-REF team should be recognized and respected by the users of the data. The TERRA-REF team reserves the right to analyze and published its own data, provided that this is done in a timely fashion. Resource users should appropriately cite the source of the data and acknowledge the resource produces. The publication the data, as suggested in the TERRA-REF Authorship Guidelines, should specify the collaborative nature of the project, and authorship is expected to include all those contributing significantly to the work.
Access to the data will be determined by the resource producers and may be governed by separate license or other agreements.
It is an objective of the TERRA-REF team to enable the release of the data to the public by November 2018 but no later than the date of close-out of the awarded funds.

The willingness of many scientists to cooperate and collaborate is what makes TERRA REF possible. Because the platform encompasses a diverse group of people and relies on many data contributors to create datasets for analysis, writing scientific papers can be more challenging than with more traditional projects. We have attempted to lay out ground rules to establish a fair process for establishing authorship, and to be inclusive while not diluting the value of authorship on a manuscript. Please engage with the TERRA REF manuscript writing process knowing you are helping to forge a new model of doing collaborative scientific research.
This document is based on the Nutrient Network Authorship Guidelines, http://nutnet.org/authorship and used with permission. Described in Borer, Elizabeth T., et al. "Finding generality in ecology: a model for globally distributed experiments."; Methods in Ecology and Evolution 5.1 (2014): 65-73.
We plan to quickly make data and software available for use with attribution, under CC-By 4.0, MIT compatable license, or Ft. Lauderdale Agreement as described in our Data Use Guidelines. Such data can be used with attribution (e.g. citation); co-authorship opportunities are welcome where warranted (see below) by specific contributions to the manuscript (e.g. help in interpreting data beyond technical support).
We are making data available early for users under the condition that manuscripts led within the team not be scooped. In these cases, people who wish to use the data for publication prior to official open release date of November 2018 should coordinate co-authorship with the person responsible for collecting the data.
Our primary goals in the TERRA REF authorship process are to consistently, accurately and transparently attribute the contribution of each author on the paper, to encourage participation in manuscripts by interested scientists, and to ensure that each author has made sufficient contribution to the paper to warrant authorship.
Steps:
Read these authorship policies and guidelines.
Consult the current list of manuscripts (http://terraref.org/manuscripts) for current proposals and active manuscripts, contact the listed lead author on any similar proposal to minimize overlap, or to join forces. Also carefully read these guidelines.
Prepare a manuscript proposal. Your proposal will list the lead author(s), the title and abstract body, and the specific data types that you will use. You can also specify more detail about response and predictor variables (if appropriate), and indicate a timeline for analysis and writing. Submit your proposal through this form.
Proposed ideas are reviewed by the authorship committee primarily to facilitate appropriate collaborations, identify potential duplication of effort, and to support the scientists who generate data while allowing the broader research community access to data as quickly and openly as possible. The authorship committee may suggest altering or combining analyses and papers to resolve issues of overlap.
Circulate your draft analysis and manuscript to solicit Opt-In authorship.
For global analyses, the lead author should circulate the manuscript to the Network by submitting a email to the TERRA REF listserv (to be determined @ terraref.org).
For analyses of more limited scope, the lead author should circulate the manuscript to network collaborators who have indicated interest at the abstract stage, those who have contributed data, and any others who the lead author deems appropriate.
In both cases, the subject line of the email should include the phrase "OPT-IN PAPER"; This email should also include a deadline by which time co-authors should respond.
The right point to share your working draft and solicit co-authors is different for each manuscript, but in general:
sharing early drafts or figures allows for more effective co-author contribution. While ideally this would mean circulating the manuscript at a very early stage for opt-in to the entire network, it is acceptable and even typical to share early drafts or figures among a smaller group of core authors.
circulating essentially complete manuscripts does not allow the opportunity for meaningful contribution from co-authors, and is discouraged.
Potential co-authors should signal their intention to opt-in by responding by email to the lead author before the stated deadline.
Lead authors should keep an email list of co-authors and communicate regularly about progress including sharing drafts of analyses, figures, and text as often as is productive and practical.
Lead authors should circulate complete drafts among co-authors and consider comments and changes. Given the wide variety of ideas and suggestions provided on each TERRA REF paper, co-authors should recognize the final decisions belong to the lead author.
Final manuscripts should be reviewed and approved by each co-author before submission.
All authors and co-authors should fill out their contribution in the authorship rubric and attach it as supplementary material to any TERRA REF manuscript. Lead authors are responsible for ensuring consistency in credit given for contributions, and may alter co-author's entries in the table to do so.
The authorship rubric provides a framework for this process.
Note that the last author position may be appropriate to assign in some cases. For example, this would be appropriate for advisors of lead authors who are graduate students or postdocs and for papers that two people worked very closely to produce.
The lead author should carefully review the authorship contribution table to ensure that all authors have contributed at a level that warrants authorship and that contributions are consistently attributed among authors. Has each author made contributions in at least two areas in the authorship rubric? Did each author provide thoughtful, detailed feedback on the manuscript? Authors are encouraged to contact the TERRA REF PI (Mockler) or authorship committee (Jeff White, Geoff Morris, Todd Mockler, David LeBauer, Wasit Wulamu, Nadia Shakoor) about any confusion or conflicts.
Authorship must be earned through a substantial contribution. Traditionally, project initiation and framing, data analysis and interpretation, and manuscript preparation are all authorship-worthy contributions, and remain so for TERRA REF manuscripts. However, TERRA REF collaborators have also agreed that collaborators who lead a site from which data are being used in a paper can also opt-in as co-authors, under the following conditions: (1) the collaborators' site has contributed data being used in the paper's analysis; and (2) that this collaborator makes additional contributions to the particular manuscript, including data analysis, writing, or editing. For coauthorship on opt-out papers, each individual must be able to check at least two boxes in the rubric, including contribution to the writing process. These guidelines apply equally to manuscripts led by graduate students.
Manuscripts published by TERRA REF will be accompanied by a supplemental table indicating authorship contributions. You can copy and share the authorship rubric. For opt-in papers, a co-author is expected to have at least two of the following areas checked in the authorship rubric.
Members: David LeBauer, Todd Mockler, Geoff Morris, Nadia Shakoor, Jeff White, Wasit Wulamu
The publications committee ensures communication across projects to avoid overlap of manuscripts, works to provide guidance on procedures and authorship guidelines, and serves as the body of last resort for resolution of authorship disputes within the Network.
Please use the following text in the acknowledgments of TERRA REF manuscripts:
The [information / data / work] presented here is from the TERRA REF experiment, funded by the Advanced Research Projects Agency-Energy (ARPA-E), U.S. Department of Energy, under Award Number DE-AR0000594. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
Please use "TERRA REF"; as one of your keywords on submitted manuscripts, so that TERRA REF work is easily indexed and searchable.
rubric item
example contribution meriting a checked box
Developed and framed research question(s)
Originated idea for current analysis of TERRA REF data; contributed significantly to framing the ideas in this analysis at early stage of manuscript
Analyzed data
Generated models (conceptual, statistical and/or mathematical), figures, tables, maps, etc.; contributed key components to the computing pipeline.
Contributed Data
generated a dataset being used in this manuscript's analysis.
Contributed to data analyses
Provided comments, suggestions, and code for data analysis
Wrote the paper
Wrote the majority of at least one of the sections of the paper
Contributed to paper writing
Provided suggestions such as restructuring ideas, text and citations linking to new literature areas, copy editing
Site level coordinator
Coordinated data collection, proofing, and submission of unreleased data for at least one site used in this manuscript.