
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
This user manual is divided into the following sections:
Data Products: A summary of the available data products and the processes used to create them
Data Access: Instructions for how to access the data products using Clowder, Globus, BETYdb, and CoGe
Description of the
: Information about data use and attribution
: In-depth examples of how to access and use the TERRA-REF data
Raw output from sensors deployed on the Lemnatec field scanner
Additional data from greenhouse systems, UAVs, and tractors have not been released, but can be accessed through our beta user program
Manually-collected fieldbooks and associated protocols
Derived data, including phenomics data, from computational approaches
You can access genomics data in one of the following locations:
Download via .
The for download or use within the CyVerse computing environment.
The computing environment.
Please review our .
The data is structured on both the TERRA-REF strorage (accessible via Globus and Workbench) and CyVerse Data Store infrastructures as follows:
Genomic pipeline data
Raw data are in bzip2 FASTQ format, one per read pair (*_R1.fastq.bz2 and *_R2.fastq.bz2). 384 samples are available. For a list of the lines sequenced, see the sample table.
Data derived from analysis of the raw resequencing data at the Danforth Center (version1) are available as gzipped, genotyped variant call format (gVCF) files and the final combined hapmap file.
Raw data are in gzip FASTQ format. 768 samples are available. For a list of lines sequenced, see the sample table.
Combined genotype calls are available in VCF format.
genomics/raw_data/ril/gbs
H5JYFBCXY_1_fastq.txt
H5JYFBCXY_2_fastq.txt
Key_ril_terra
genomics/derived_data/ril/gbs/kansas_state/version1/imp_TERRA_RIL_SNP.vcf
|-terraref
| |-genomics
| | |-raw_data
| | | |-bap
| | | | |-resequencing
| | | |-ril
| | | | |-gbs
| | |-derived_data
| | | |-bap
| | | | |-resequencing
| | | | | |-danforth_center
| | | |-ril
| | | | |-gbs
| | | | | |-kansas_stateThe following table lists available TERRA-REF data products. The table will be updated as new datasets are released. Links are provided to pages with detailed information about each data product including sensor descriptions, algorithm (extractor) information, protocols, and data access instructions.
Multispectral data is collected using the PRI and NDVI Skye sensors. Raw output is converted to timeseries data using the multispectral extractor.
Stereo imaging data is collected using the Prosilica cameras. Full-color images are reconstructed in GeoTIFF format using the de-mosaic extractor. A full-field mosaic is generated using the full-field mosaic extractor.
Spectral reflectance data
Spectral reflectance is measured using a Crop Circle active crop canopy sensor
Environment conditions are collected through the CO2 sensor and Thies Clima. Raw output is converted to netCFG using the environmental-logger extractor.
postGIS/netCDF
Phenotype data is derived from sensor output using the PlantCV extractor and imported into BETYdb.
FASTQ and VCF files available via Globus
UAV and Phenotractor
Plot level data available in BETYdb
Data product
Description
3D point cloud data (LAS) of the field constructed from the Fraunhofer 3D scanner output (PLY).
Fluorescence intensity imaging is collected using the PSII LemnaTec camera. Raw camera output is converted to (netCDF/GeoTIFF)
Hyperspectral imaging data from the SWIR and VNIR Headwall Inspector sensors are converted to netCDF output using the hyperspectral extractor.
Infrared heat imaging data is collected using FLIR sensor. Raw output is converted to GeoTIFF using the FLIR extractor.
Several different sensors include geospatial information in the dataset metadata describing the location of the sensor at the time of capture.
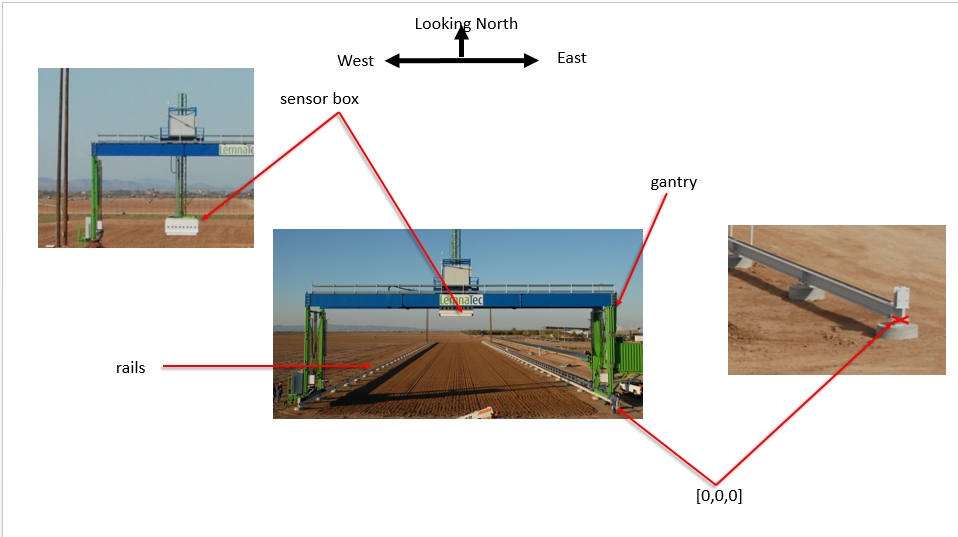
Coordinate reference systems The Scanalyzer system itself does not have a reliable GPS unit on the sensor box. There are 3 different coordinate systems that occur in the data:
Most common is EPSG:4326 (WGS84) USDA coordinates
Tractor planting & sensor data is in UTM Zone 12
Sensor position information is captured relative to the southeast corner of the Scanalyzer system in meters
ua-mac/Level_1/ir_geotiff
Plot level summaries are named 'surface_temperature' in the trait database. In the future this name will be used for the Level 1 data as well. This name from the Climate Forecast (CF) conventions, and is used instead of 'canopy_temperature' for two reasons: First, because we do not (currently) filter soil in this pipeline. Second, because the CF definition of surface_temperature distinguishes the surface from the medium: "The surface temperature is the temperature at the interface, not the bulk temperature of the medium above or below." http://cfconventions.org/Data/cf-standard-names/48/build/cf-standard-name-table.html
Thermal imaging data is available via Clowder and Globus:
/ua-mac/raw_data/flirIrCamera
For details about using this data via Clowder or Globus, please see Data Access section.
Data are unavailable for Season 4 (summer 2017 sorghum) and season 5 (winter 2017-2018 wheat).
Work to recover these data is ongoing; see terraref/reference-data#190
Problem description terraref/reference-data#182
EPSG:4326 coordinates for the four corners of the Scanalyzer system (bound by the rails above) are as follows:
NW: 33° 04.592' N, -111° 58.505' W
NE: 33° 04.591' N, -111° 58.487' W
SW: 33° 04.474' N, -111° 58.505' W
SE: 33° 04.470' N, -111° 58.485' W
In the trait database, this site is named the "MAC Field Scanner Field" and its bounding polygon is "POLYGON ((-111.9747967 33.0764953 358.682, -111.9747966 33.0745228 358.675, -111.9750963 33.074485715 358.62, -111.9750964 33.0764584 358.638, -111.9747967 33.0764953 358.682))"
Scanalyzer coordinates Finally, the Scanalyzer coordinate system is right-handed - the origin is in the SE corner, X increases going from south to north, and Y increases from east to the west.
In offset meter measurements from the southeast corner of the Scanalyzer system, the extent of possible motion for the sensor box is defined as:
NW: (207.3, 22.135, 5.5)
SE: (3.8, 0, 0)
Scanalyzer -> EPSG:4326 1. Calculate the UTM position of known SE corner point 2. Calculate the UTM position of the target point, using SE point as reference 3. Get EPSG:4326 position based on UTM
MAC coordinates Tractor planting data and tractor sensor data will use UTM Zone 12.
Scanalyzer -> MAC Given a Scanalyzer(x,y), the MAC(x,y) in UTM zone 12 is calculated using the linear transformation formula:
Assume Gx = -Gx', where Gx' is the Scanalyzer X coordinate.
MAC -> Scanalyzer
MAC -> EPSG:4326 USDA We do a linear shifting to convert MAC coordinates in to EPSG:4326 USDA
Sensors with geospatial metadata
stereoTop
flirIr
co2
cropCircle
PRI
scanner3dTop
NDVI
PS2
SWIR
VNIR
Available data All listed sensors
stereoTop
cropCircle
co2Sensor
flirIrCamera
ndviSensor
priSensor
SWIR
field scanner plots
There are 864 (54*16) plots in total and the plot layout is described in the plot plan table.
dimension
value
# rows
32
# rows / plot
2
# plots (2 rows ea)
864
# ranges
54
# columns
16
The boundary of each plot changes slightly each planting season. The scanalyzer coordinates of each plot are transformed into the (EPSG:4326) USDA coordinates using the equations above.
3D point cloud data is collected using the Fraunhofer 3D laser scanner. Custom software installed at Maricopa converts .png output to the .ply point clouds. The .ply point clouds are converted to georeferenced .las files using the 3D point cloud extractor
Level 1 data products are provided in both .las and .ply formats. Raw sensor output (PLY) is converted to LAS format using the ply2las extractor; ply2las extractor code is available on GitHub.
For each scan, there are two .ply files representing two lasers, one on the left and the other on the right. These are combined in the .las files.
For details about using this data via Clowder or Globus, please see section.
Data is available via Clowder, Globus, and Workbench.
Clowder:
Globus or Workbench File System:
LAS /raw_data/laser3D_las
PLY /raw_data/laser3D
The position of the lasers is affected by temperature. We added a correction for temperature to adjust for this effect. See
TERRA-REF data can be accessed through many different interfaces: Globus, Clowder, BETYdb, CyVerse, and CoGe. Raw data is transfered to the primary compute pipeline using Globus Online. Data is ingested into Clowder to support exploratory analysis. The Clowder extractor system is used to transform the data and create derived data products, which are either available via Clowder or published to specialized services, such as BETYdb.
ay = 3659974.971; by = 1.0002; cy = 0.0078;
ax = 409012.2032; bx = 0.009; cx = - 0.9986;
Mx = ax + bx * Gx + cx * Gy
My = ay + by * Gx + cy * GyGx = ( (My/cy - ay/cy) - (Mx/cx - ax/cx) ) / (by/cy - bx/cx)
Gy = ( (My/by - ay/by) - (Mx/bx - ax/bx) ) / (cy/by - cx/bx)Latitude: Uy = My - 0.000015258894
Longitude: Ux = Mx + 0.000020308287"gantry_system_variable_metadata": {
"time": "08/17/2016 11:23:14",
"position x [m]": "207.013",
"position y [m]": "3.003",
"position z [m]": "0.68",
"speed x [m/s]": "0",
"speed y [m/s]": "0.33",
"speed z [m/s]": "0",
"camera box light 1 is on": "True",
"camera box light 2 is on": "True",
"camera box light 3 is on": "True",
"camera box light 4 is on": "True",
"y end pos [m]": "22.135",
"y set velocity [m/s]": "0.33",
"y set acceleration [m/s^2]": "0.1",
"y set decceleration [m/s^2]": "0.1"
},"sensor_fixed_metadata": {
"cameras alignment": "cameras optical axis parallel to XAxis, perpendicular to ground",
"optics focus setting (both)": "2.5m",
"optics apperture setting (both)": "6.7",
"location in gantry system": "camera box, facing ground",
"location in camera box x [m]": "0.877",
"location in camera box y [m]": "2.276",
"location in camera box z [m]": "0.578",
"field of view at 2m in X- Y- direction [m]": "[1.857 1.246]",
"bounding Box [m]": "[1.857 1.246]",
},"sensor_fixed_metadata": {
"location in gantry system": "camera box, facing ground",
"location in camera box x [m]": "0.480",
"location in camera box y [m]": "1.920",
"location in camera box z [m]": "0.6",
},"sensor_fixed_metadata": {
"location in gantry system": "camera box, facing ground",
"location in camera box x [m]": "0.35",
"location in camera box y [m]": "2.62",
"location in camera box z [m]": "0.7",
},"sensor_fixed_metadata": {
"location in gantry system": "camera box, facing ground",
"location in camera box x [m]": "0.877",
"location in camera box y [m]": "1.361",
"location in camera box z [m]": "0.520",
"field of view x [m]": "1.496",
"field of view y [m]": "1.105",
},"sensor_fixed_metadata": {
"location in gantry system": "top of gantry, facing up, camera box, facing ground",
"location in camera box x [m]": "0.33",
"location in camera box y [m]": "2.50",
},"sensor_fixed_metadata": {
"location in gantry system": "top of gantry, facing up, camera box, facing ground",
"location in camera box x [m]": "0.400",
"location in camera box y [m]": "2.470",
},"sensor_fixed_metadata": {
"location in gantry system": "camera box, facing ground",
"location in camera box x [m]": "0.877",
"location in camera box y [m]": "2.325",
"location in camera box z [m]": "0.635",
"field of view y [m]": "0.75",
"optics focal length [mm]": "25",
"optics focus apperture": "2.0",
},clients
Sensor Data
Globus
Browse directories; transfer large sensor files
globus.org #TERRAREF endpoint
R, Python
Clowder
Browse and Download small Sensor Data
terraref.org/clowder
Python
Trait Data
BETYdb
Trait and Agronomic Metadata
terraref.org/bety
and
R traits package, Python: terrautils; SQL: Postgres in Docker
traitvis
View available trait data
terraref.org/traitvis
NA
NA
Genomics Data
CyVerse
Download Genomics data
terraref.org/cyverse-genomics
yes
CoGe
Download, process, visualize Genomics data
terraref.org/coge
Other
Tutorials
R and Python scripts for accessing data
terraref.org/tutorials
NA
Advanced Search
Search across sensor and trait data
search.terraref.org (under development)
yes
We have developed tutorials to provide users with both 'quick start' vignettes and more detailed introductions to TERRA REF datasets. Tutorials for accessing trait data, sensor data, and genomics data are organized by directory ("traits", "sensors", and "genomics").
The tutorials assume familiarity with or willingness to learn Python and / or R, and provide the greatest flexibility and access to available data.
These can be found at terraref.org/tutorials.
Raw data is transferred to the primary TERRA-REF file system at the National Center for Computing Applications at the University of Illinois.
Public domain data is available for Globus transfer via the ncsa#terra-public. Non-public (but available with permission) data are at the #Terraref endpoint
Use Globus Online when you want to transfer data from the TERRA-REF system for local analysis.
See also Globus Getting Started
The Globus Connect service provides high-performance, secure, file transfer and synchronization between endpoints. It also allows you to securely share your data with other Globus users.
To access data via Globus, you must first have a Globus account and endpoint.
Sign up for Globus at globus.org
Log into Globus https://www.globus.org
Add an endpoint for the destination (e.g. your local computer)
Go to the 'transfer files' page:
Select source
Endpoint: #Terraref
Path: Navigate to the subdirectory that you want.
Select (click) a folder
Click 'go'
Files will be transfered to your computer
Requesting Access to unpublished data in TERRA-REF BETYdb:
To request access to unpublished data, send your Globus id to David LeBauer ([email protected]) with 'TERRAREF Globus Access Request' in the subject.
fill out the terraref.org/beta user form
email [email protected] with your globusid to request access.
BETYdb is used to manage and distribute agricultural and ecological data. It contains phenotype and agronomic data including plot locations and other geolocations of interest (e.g. fields, rows, plants).
BETYdb contains the derived trait data with plot locations and other information associated with agronomic experimental design.
The easiest way to access data is to use the R traits package. This is documented in the tutorials.
Requesting Access to unpublished data in TERRA-REF BETYdb:
fill out the terraref.org/beta user form
create an account at the TERRA-REF BETYdb: terraref.org/bety (not betydb.org)
email [email protected] for your account to be approved.
The fastest and most comprehensive way to access the database using SQL and other database interfaces (such as the R package dplyr interface described below, or GIS programs described in . You can run an instance of the database using docker, as described below
This is how you can access the TERRA REF trait database. It requires that you install the Docker software on your computer.
The easiest way to get the entire database, including metadata. Assuming you are familiar with the Postgres and / or the R dbplyr library documentation. See the TERRA REF Tutorials terraref.org/tutorials, the BETYdb Data Access guide for additional examples.
Interested researchers can access BETYdb directly from GIS software such as ESRI ArcMap and QGIS. In some cases direct access can simplify the use of spatial data in BETYdb. See the Appendix Accessing BETYdb with GIS Software for more information.
Clowder is an active data repository designed to enable collaboration around a set of shared datasets. TERRAREF uses Clowder to organize, annotate, and process data generated by phenotyping platforms. Datafiles are available via the Clowder web interface or API.
Clowder is the used to organize, annotate, and process raw data generated by the field scanner and other phenotyping platforms. It also stores information about sensors. Learn more about Clowder software from https://clowderframework.org
Data is organized into spaces, collections, and datasets, collections.
Spaces contain collections and datasets. TERRA-REF uses one space for each of the phenotyping platforms.
Collections consist of one or more datasets. TERRA-REF collections are organized by acquisition date and sensor. Users can also create their own collections.
Datasets consist of one or more files with associated metadata collected by one sensor at one time point. Users can annotate, download, and use these sensor datasets.
Requesting Access to unpublished data in Clowder:
fill out the terraref.org/beta user form
create an account at the TERRA-REF Clowder site
email [email protected] for your account to be approved.
CyVerse is a National Science Foundation funded cyberinfrastructure that aims to democratize access to supercomputing capabilities.
TERRA-REF genomics data is accessible on the CyVerse Data Store and Discovery Environment. Accessing data through the CyVerse Discovery Environment requires signing up for a free CyVerse account. The Discovery Environment gives users access to software and computing resources, so this method has the advantage that TERRA-REF data can be utilized directly without the need to copy the data elsewhere.
Genomics data can be browsed and downloaded from the CyVerse data store at http://datacommons.cyverse.org/browse/iplant/home/shared/terraref
You can also find these in the CyVerse discovery environment in the TERRA-REF Community Data folder: /iplant/home/shared/terraref.
CoGe is a platform for performing Comparative Genomics research. It provides an open-ended network of interconnected tools to manage, analyze, and visualize next-gen data.
CoGe contains genomic information and sequence data. You can find the TERRA REF Genomics data on CoGe in this notebook: https://genomevolution.org/coge/NotebookView.pl?nid=2137
Resource
Use
Web User Interface
API*
row width (m)
0.762
plot length (m)
4
row length (m)
3.5
alley length (m)
0.5
#git clone https://github.com/terraref/data-paper
cd data-paper/code/betydb_docker
docker-compose up -d postgres
docker-compose run --rm bety initialize
docker-compose run --rm bety syncpsql -d bety -U bety -W betylibrary(dplyr)
bety_src <- src_postgres(dbname = "bety",
password = 'bety',
host = 'localhost',
user = 'bety',
port = 5433)Select (highlight) files that you want to download at destination
Select the endpoint that you set up above of your local computer or server
Select the destination folder (e.g. /~/Downloads/)
Data will be released with a CC0 license, meaning that they are in the public domain. The CC0 license allows wide use of these data and while it does not legally bind users to acknowledge the source data users are expected cite our data and research in publications, presentations, and other products.
The first release in 2020 included data from Seasons 4 and 6. This is available on Dryad and Globus.
Data are in the public domain to enable broad and unrestricted re-use. However, any derived pubications should cite the dataset:
Genomics, sensor, and phenotype data: LeBauer, David et al. (2020), Data From: TERRA-REF, An open reference data set from high resolution genomics, phenomics, and imaging sensors, Dryad, Dataset,
AZMet Weather Data (available on Dryad): Brown, P. W., and B. Russell. 1996. “AZMET, the Arizona Meteorological Network. Arizona Cooperative Extension.” .
Data Processing Pipeline: Burnette, Maxwell, et al. "TERRA-REF data processing infrastructure." Proceedings of the Practice and Experience on Advanced Research Computing. 2018. 1-7.
For algorithms, we intend to release via BSD 3 clause or MIT / BSD compatible license. Algorithms are available on GitHub in the terraref organization: and have been archived on Zenodo (see ).
Citations for Individual Software and Documentation Components are listed in the section and can be browsed on .
We plan to make data from the Transportation Energy Resources from Renewable Agriculture Phenotyping Reference Platform (TERRA-REF) project available for use with attribution.
Please consider engaging with team members to collaborate on new research with these data. You can learn more about our approach to co-authorship and also about planned research papers in the section .
Any access to data prior to publication is granted with the understanding that the contributions and interests of the TERRA-REF team should be recognized and respected by the users of the data. The TERRA-REF team reserves the right to analyze and published its own data. Resource users should appropriately cite the source of the data and acknowledge the resource produces. The publication of the data, as suggested in the , should specify the collaborative nature of the project, and authorship is expected to include all those TERRA-REF team members contributing significantly to the work.
Planned future releases include Sorghum Season 9, data from experiments at Kansas State University, and data from the Danforth Indoor Phenotyping Facility.
Additional seasons can be requested as needed. We can provide the raw data and software required to process it. We can also collaborate with you to process the data, but this will typically require new funding sources.
Todd Mockler, Project/Genomics Lead (email: [email protected])
David LeBauer, Computing Pipeline Lead (email: [email protected])
Nadia Shakoor, Project Director (email: [email protected])
See also in the Introduction.
Fluorescence intensity data is collected using the PSII camera.
Each measurement produces 102 bin files. The first (index 0) is an image taken right before the LED are switched on (dark reference). Frame 1 to 100 are the 100 images taken, with the LEDs on. In binary file 102 (index 101) is a list with the timestamps of each frame of the 100 frames.
Right now the LED on timespan is 1s thus the first 50 frames are taken with LEDs on the latter 50 frames with LED off.
Fluorescence intensity data is available via Clowder and Globus:
Clowder: __
Globus paths:
/raw_data/ps2top
Herritt, Matthew T., et al. "Chlorophyll fluorescence imaging captures photochemical efficiency of grain sorghum (Sorghum bicolor) in a field setting." Plant Methods 16.1 (2020): 1-13.
/Level_1/ps2_png/Sensor information: LemnaTec PSII
Code on GitHub: Multispectral extractor
Environment conditions data is collected using the Vaisala CO2, Thies Clima weather sensors as well as lightning, irrigation, and weather data collected at the Maricopa site.
Data formats follow the Climate and Forecast (CF) conventions for variable names and units. Environmental data are stored in the Geostreams database.
WeatherStation coordinates are 33.074457 N, 111.975163 W
EnvironmentLogger is on top of the gantry system and is moveable.
Irrigation is managed at the field level. There are four regions that can be irrigated at different rates.
Level 1 meteorological data is aggregated to from 1 Hz raw data to 5 minute averages or sums.
On Globus or Workbench you can find these data provided in both hourly and daily files. These files contain data at the original temporal resolution of 1/s. In addition, they contain the high resolution spectral radiometer data.
sites/ua-mac/Level_1/envlog_netcdf
hourly files: YYYY-MM-DD_HH-MM-SS_environmentallogger.nc
daily files: envlog_netcdf_L1_ua-mac_YYYY-MM-DD.nc
Data can be accessed using the geostreams API or the PEcAn meteorological workflow. These are illustrated in the .
Here is the json representation of a single five-minute observation:
Data can be accessed using the geostreams API or the PEcAn meteorological workflow.
These are illustrated in the .
Here is the json representation of a single five-minute observation from Geostreams:
Data is available via Globus or Workbench:
/ua-mac/raw_data/co2sensor
/ua-mac/raw_data/EnvironmentLogger
/ua-mac/raw_data/irrigation
/ua-mac/raw_data/lightning
Description: EnvironmentalLogger raw files are converted to netCDF.
Known issue: the irrigation data stream does not currently handle variable irrigation rates within the field. Specifically, we have not yet accounted for the Summer 2017 drought experiments. See for more information.
When the full field is irrigated (as is typical), the irrigated area is 5466.1 m2 (=215.2 m x 25.4 m)
In 2017:
Full field irrigated area from the start of the season to August 1 (103 dap) is 5466.1 m2 (=215.2 m x 25.4 m).
Well-watered treatment zones from August 1 to 15 (103 to 116 dap): 2513.5 m2 (=215.2 m x 11.68 m) in total, combined areas of non-contiguous blocks
Well-watered treatment zones from August 15 - 30 (116 to 131 dap): 3169.9 m2 (=215.2 m x 14.73 m), again in total as the combined areas of non-contiguous blocks
Phenotype data is derived from images generated by the indoor LemnaTec Scanalyzer 3D platform at the Donald Danforth Plant Science Center using PlantCV. PlantCV is an image analysis package for plant phenotyping. PlantCV is composed of modular functions in order to be applicable to a variety of plant types and imaging systems. PlantCV contains base functions that are required to examine images from an excitation imaging fluorometer (PSII), visible spectrum camera (VIS), and near-infrared camera (NIR). PlantCV is a fully open source project: https://github.com/danforthcenter/plantcv. For more information, see:
Project website: http://plantcv.danforthcenter.org
Full documentation:
Publications:
To learn more about PlantCV, you can find examples in the repository, which is accessible on GitHub and in the TERRA REF under tutorials/plantcv
an ipython notebook demonstration of PlantCV .
For the TERRA-REF project, a PlantCV Clowder extractor was developed to analyze data from the at the Donald Danforth Plant Science Center. Resulting phenotype data is stored in BETYdb.
Description: Processes VIS/NIR images captured at several angles to generate trait metadata. The trait metadata is associated with the source images in Clowder, and uploaded to the configured BETYdb instance.
Output CSV: /sites/danforth/Level_1/<experiment name>
Input
Evaluation is triggered whenever a file is added to a dataset
Following images must be found
2x NIR side-view = NIR_SV_0, NIR_SV_90
1x NIR top-view = NIR_TV
Output
Each image will have new metadata appended in Clowder including measures like height, area, perimeter, and longest_axis
Average traits for the dataset (3 VIS or 3 NIR images) are inserted into a CSV file and added to the Clowder dataset
If configured, the CSV will also be sent to BETYdb
BETYdb:
For details about accessing BETYdb, please see section and a tutorial on accessing phenotypes from the trait database on the TERRA REF Workbench in .
Clowder:
Globus and Workbench:
/sites/danforth/raw_data/<experiment name>
Meteorological data will use conventions. CF is widely used in climate, meteorology, and earth sciences.
Here are some examples (note that we can change from canonical units to match the appropriate scale, e.g. "C" instead of "K"; time can use any base time and time step (e.g. hours since 2015-01-01 00:00:00 UTC, etc. But the time zone has to be UTC, where 12:00:00 is approx (+/- 15 min). solar noon at Greenwich.
2x VIS side-view = VIS_SV_0, VIS_SV_90
1x VIS top-view = VIS_TV
Per-image metadata in Clowder is required for BETYdb submission; this is how barcode/genotype/treatment/timestamp are determined.
K
tasmaxAdjust
NA
tmax
air_temperature_min
K
tasminAdjust
NA
tmin
air_pressure
Pa
air_pressure
PRESS (KPa)
mole_fraction_of_carbon_dioxide_in_air
mol/mol
CO2
moisture_content_of_soil_layer
kg m-2
soil_temperature
K
soilT
TS1 (NOT DONE)
relative_humidity
%
relative_humidity
rhurs
NA
rhum
RH
specific_humidity
1
specific_humidity
NA
qair
shum
CALC(RH)
water_vapor_saturation_deficit
Pa
VPD
VPD (NOT DONE)
surface_downwelling_longwave_flux_in_air
W m-2
same
rldsAdjust
lwdown
dlwrf
Rgl
surface_downwelling_shortwave_flux_in_air
W m-2
solar_radiation
rsdsAdjust
swdown
dswrf
Rg
surface_downwelling_photosynthetic_photon_flux_in_air
mol m-2 s-1
PAR
PAR (NOT DONE)
precipitation_flux
kg m-2 s-1
cccc
prAdjust
rain
acpc
PREC (mm/s)
degrees
wind_direction
WD
wind_speed
m/s
Wspd
WS
eastward_wind
m/s
eastward_wind
CALC(WS+WD)
northward_wind
m/s
northward_wind
CALC(WS+WD)
/ua-mac/raw_data/weather
CF standard-name
units
bety
isimip
cruncep
narr
ameriflux
air_temperature
K
airT
tasAdjust
tair
air
TA (C)
air_temperature_max
Pa
mole_fraction_of_carbon_dioxide_in_air
mol/mol
moisture_content_of_soil_layer
kg m-2
soil_temperature
K
relative_humidity
%
specific_humidity
1
water_vapor_saturation_deficit
Pa
surface_downwelling_longwave_flux_in_air
W m-2
surface_downwelling_shortwave_flux_in_air
W m-2
surface_downwelling_photosynthetic_photon_flux_in_air
mol m-2 s-1
precipitation_flux
kg m-2 s-1
irrigation_flux
kg m-2 s-1
irrigation_transport
kg s-1
wind_speed
m/s
eastward_wind
m/s
northward_wind
m/s
standard_name is CF-convention standard names (except irrigation)
units can be converted by udunits, so these can vary (e.g. the time denominator may change with time frequency of inputs)
Before the Running
The pipepline is developed in Python, so a Python Interpreter is a must. Other than the basic Python standard librarys, the following third-party libraries are required:
netCDF4 for Python
numpy
Other than official CPython interpreter, Pypy is also welcomed, but please make sure that these third-party modules are correctly installed for the target interpreter. The pipeline can only works in Python 2.X versions (2.7 recommended) since numpy does not support Python 3.X versions.
Cloning from the Git:
The extractor for this pipeline is developed and maintained by Max in branch "EnvironmentalLogger-extractor" under the same repository.
Get the Environmental Logger Pipeline to Work
To trigger the pipeline, use the following command:
python ${environmental_logger_source_path}/environmental_logger_json2netcdf.py ${input_JSON_file} ${output_netCDF_file}
Where:
${environmental_logger_source_path} is where the three environmental_logger files are located
${input_JSON_file} is where the input JSON files are located
${output_netCDF_file} is where the users want the pipeline to export the product (netCDF file)
Please note that the parameter for the output file can be a path to either a directory or a file, and it is not necessarily to be existed. If the output is a path to a folder, the final product will be in this folder as a netCDF file that has the same name as the imported JSON file but with a different filename extension (.nc for standard netCDF file); if this path does not exist, environmental_logger pipeline will automatically make one.
The calculation in the Environmental Logger is mainly finished by the module environmental_logger_calculation.py under the support of numpy.
CF standard-name
units
time
days since 1700-01-01 00:00:00 UTC
air_temperature
K
air_pressure
[
{
"geometry":{
"type":"Point",
"coordinates":[
33.0745666667,
-111.9750833333,
0
]
},
"start_time":"2016-08-30T00:06:24-07:00",
"type":"Feature",
"end_time":"2016-08-30T00:10:00-07:00",
"properties":{
"precipitation_rate":0.0,
"wind_speed":1.6207870370370374,
"surface_downwelling_shortwave_flux_in_air":0.0,
"northward_wind":0.07488770951583902,
"relative_humidity":26.18560185185185,
"air_temperature":300.17606481481516,
"eastward_wind":1.571286062845733,
"surface_downwelling_photosynthetic_photon_flux_in_air":0.0
}
},git clone https://github.com/terraref/computing-pipeline.git
cd computing-pipeline/scripts/environmental_logger
git checkout master






The willingness of many scientists to cooperate and collaborate is what makes TERRA REF possible. Because the platform encompasses a diverse group of people and relies on many data contributors to create datasets for analysis, writing scientific papers can be more challenging than with more traditional projects. We have attempted to lay out ground rules to establish a fair process for establishing authorship, and to be inclusive while not diluting the value of authorship on a manuscript. Please engage with the TERRA REF manuscript writing process knowing you are helping to forge a new model of doing collaborative scientific research.
This document is based on the Nutrient Network Authorship Guidelines, http://nutnet.org/authorship and used with permission. Described in Borer, Elizabeth T., et al. "Finding generality in ecology: a model for globally distributed experiments."; Methods in Ecology and Evolution 5.1 (2014): 65-73.
We plan to quickly make data and software available for use with attribution, under , compatable license, or Ft. Lauderdale Agreement as described in our . Such data can be used with attribution (e.g. citation); co-authorship opportunities are welcome where warranted (see below) by specific contributions to the manuscript (e.g. help in interpreting data beyond technical support).
We are making data available early for users under the condition that manuscripts led within the team not be scooped. In these cases, people who wish to use the data for publication prior to official open release date of November 2018 should coordinate co-authorship with the person responsible for collecting the data.
Our primary goals in the TERRA REF authorship process are to consistently, accurately and transparently attribute the contribution of each author on the paper, to encourage participation in manuscripts by interested scientists, and to ensure that each author has made sufficient contribution to the paper to warrant authorship.
Steps:
Read these authorship policies and guidelines.
Consult the current list of manuscripts () for current proposals and active manuscripts, contact the listed lead author on any similar proposal to minimize overlap, or to join forces. Also carefully read these guidelines.
Prepare a manuscript proposal. Your proposal will list the lead author(s), the title and abstract body, and the specific data types that you will use. You can also specify more detail about response and predictor variables (if appropriate), and indicate a timeline for analysis and writing. Submit your proposal through .
Proposed ideas are reviewed by the authorship committee primarily to facilitate appropriate collaborations, identify potential duplication of effort, and to support the scientists who generate data while allowing the broader research community access to data as quickly and openly as possible. The authorship committee may suggest altering or combining analyses and papers to resolve issues of overlap.
Authors are encouraged to contact the TERRA REF PI (Mockler) or authorship committee (Jeff White, Geoff Morris, Todd Mockler, David LeBauer, Wasit Wulamu, Nadia Shakoor) about any confusion or conflicts.
Authorship must be earned through a substantial contribution. Traditionally, project initiation and framing, data analysis and interpretation, software or algorithm development, and manuscript preparation are all authorship-worthy contributions, and remain so for TERRA REF manuscripts. However, TERRA REF collaborators have also agreed that collaborators who lead a site from which data are being used in a paper can also opt-in as co-authors, under the following conditions: (1) the collaborators' site has contributed data being used in the paper's analysis; and (2) that this collaborator makes additional contributions to the particular manuscript, including data analysis, writing, or editing. For co-authorship on opt-out papers, each individual must be able to check at least two boxes in the rubric in addition to contribution to the writing process. These guidelines apply equally to manuscripts led by graduate students.
This section is derived from the International Committee of Medical Journal Editors (ICMJE) .
Each author is expected to meet all of the following conditions:
Substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data, and
Drafting the article or revising it critically for important intellectual content, and
Final approval of the version to be published, and
Manuscripts published by TERRA REF will be accompanied by a supplemental table indicating authorship contributions. You can copy and share the . For opt-in papers, a co-author usually should contribute to writing and revision as well as at least two of the following areas checked in the authorship rubric. This follows the CRediT Taxonomy as published in the .
By default, we will follow the conventions of the scientific community that is the target audience of the journal in which the article is published. This should typically follow:
First is lead author
Last is the supervisor of the lead author.
if > 1 lead or senior authors these will be listed first and last, respectively, and identified in the author contributions section of the acknowledgements.
All other contributors are listed alphabetically.
Members: David LeBauer, Todd Mockler, Geoff Morris, Duke Pauli, Nadia Shakoor, Wasit Wulamu
The publications committee ensures communication across projects to avoid overlap of manuscripts, works to provide guidance on procedures and authorship guidelines, and serves as the body of last resort for resolution of authorship disputes within the Network.
Please use the following text in the acknowledgments of TERRA REF manuscripts:
The [information / data / work] presented here is from the experiment, funded by the Advanced Research Projects Agency-Energy (ARPA-E), U.S. Department of Energy, under Award Number DE-AR0000594. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
Please use "TERRA REF"; as one of your keywords on submitted manuscripts, so that TERRA REF work is easily indexed and searchable.
Circulate your draft analysis and manuscript to solicit Opt-In authorship.
For global analyses, the lead author should circulate the manuscript to the Network by submitting a email to the TERRA REF team.
For analyses of more limited scope, the lead author should circulate the manuscript to network collaborators who have indicated interest at the abstract stage, those who have contributed data, and any others who the lead author deems appropriate.
In both cases, the subject line of the email should include the phrase "OPT-IN PAPER"; This email should also include a deadline by which time co-authors should respond.
The right point to share your working draft and solicit co-authors is different for each manuscript, but in general:
sharing early drafts or figures allows for more effective co-author contribution. While ideally this would mean circulating the manuscript at a very early stage for opt-in to the entire network, it is acceptable and even typical to share early drafts or figures among a smaller group of core authors.
circulating essentially complete manuscripts does not allow the opportunity for meaningful contribution from co-authors, and is discouraged.
Potential co-authors should signal their intention to opt-in by responding by email to the lead author before the stated deadline.
Potential co-authors should inform the lead author of any additional candidates for co-authorship who should be considered.
Lead authors should is responsible for making sure that any who have made contributions warranting co-authorship have actively opted in or out (authors should not be excluded due to a missed email or a misunderstanding of the scope of the manuscript and their contributions). The goal is to ensure that the author list is inclusive and consistent.
Lead authors should keep an email list of co-authors and communicate regularly about progress including sharing drafts of analyses, figures, and text as often as is productive and practical.
Lead authors should circulate complete drafts among co-authors and consider comments and changes. Given the wide variety of ideas and suggestions provided on each TERRA REF paper, co-authors should recognize the final decisions belong to the lead author.
Final manuscripts should be reviewed and approved by each co-author before submission.
All authors and co-authors should fill out their contribution in the authorship rubric and attach it as supplementary material to any TERRA REF manuscript. Lead authors are responsible for ensuring consistency in credit given for contributions, and may alter co-author's entries in the table to do so.
The authorship rubric provides a framework for the opt-in process. Lead authors should copy the template and edit the contents for a specific manuscript, then circulate to potential co-authors.
Note that the last author position may be appropriate to assign in some cases. For example, this would be appropriate for advisors of lead authors who are graduate students or postdocs and for papers that two people worked very closely to produce.
The lead author should carefully review the authorship contribution table to ensure that all authors have contributed at a level that warrants authorship and that contributions are consistently attributed among authors. The lead author should also ensure that all contributions that warrant co-authorship.
Has each author made contributions in at least two areas in the authorship rubric?
Did each author provide thoughtful, detailed feedback on the manuscript?
Have all qualified contributors actively opted in or out of co-authorship?
Development or design of methodology; creation of models
Project Administration
Management and coordination responsibility for the research activity planning and execution.
Resources
Provision of study materials, reagents, materials, patients, laboratory samples, animals, instrumentation, computing resources, or other analysis tools.
Software
Programming, software development; designing computer programs; implementation of the computer code and supporting algorithms; testing of existing code components.
Supervision
Oversight and leadership responsibility for the research activity planning and execution, including mentorship external to the core team.
Validation
Verification, whether as a part of the activity or separate, of the overall replication/reproducibility of results/experiments and other research outputs.
Visualization
Preparation, creation and/or presentation of the published work, specifically visualization/data presentation.
Writing – Original Draft Preparation
Creation and/or presentation of the published work, specifically writing the initial draft (including substantive translation).
Writing – Review & Editing
Preparation, creation and/or presentation of the published work by those from the original research group, specifically critical review, commentary or revision – including pre- or post-publication stages.
Contributor Role
Role Definition
Conceptualization
Ideas; formulation or evolution of overarching research goals and aims.
Data Curation
Management activities to annotate (produce metadata), scrub data and maintain research data (including software code, where it is necessary for interpreting the data itself) for initial use and later reuse.
Formal Analysis
Application of statistical, mathematical, computational, or other formal techniques to analyze or synthesize study data.
Funding Acquisition
Acquisition of the financial support for the project leading to this publication.
Investigation
Conducting a research and investigation process, specifically performing the experiments, or data/evidence collection.
Methodology


Hyperspectral imaging data is collected using the Headwall VNIR and SWIR sensors. In the Nov 2017 Beta Release only VNIR data is provided because we do not have the measurements of downwelling spectral radiation required by the pipeline.
Please see the README hyperspectral pipeline README for more information about how the data are generated and known issues.
See
Raw data is available in the filesystem, accessible via Globus in the following directories:
VNIR: /sites/ua-mac/raw_data/VNIR
SWIR: /sites/ua-mac/raw_data/SWIR
These files are uncalibrated; see the hyperspectral pipeline repository for information on how these can be processed.
Hyperspectral data is available via Clowder, , the , and our :
Clowder:
SWIR Collection: Level 1 data not available
Globus and Workbench:
For details about using this data via Clowder or Globus, please see section.
Level 2 data are spectral indices computed at the same resolution as Level 1. These can be found in the same Level 1 directories as their parents, but the files are appended *_ind.nc.
To get a list of hyperspectral indices currently generated
The following indices are computed and provided as both Level 2 data at full spatial resolution and as Level 3 (plot level) means.
Citations can be found Morris, Geoffrey P., Davina H. Rhodes, Zachary Brenton, Punna Ramu, Vinayan Madhumal Thayil, Santosh Deshpande, C. Thomas Hash et al. "Dissecting genome-wide association signals for loss-of-function phenotypes in sorghum flavonoid pigmentation traits." G3: Genes, Genomes, Genetics 3, no. 11 (2013): 2085-2094.
VNIR: /sites/ua-mac/Level_1/vnir_netcdf
SWIR: Level 1 data not available
Sensor information:
R900/R680
Rouse et al. (1973)
DWSI2
Disease Water Stress Index 2
R1660 / R550
Apan, Held, Phinn and Markley (2003)
TCARI
Transformed Chlorophyll Absorption Ratio
3 ((R700 - R670) - 0.2 (R700 - R550) * (R700/R670))
Haboudane et al. (2002)
DWSI3
Disease Water Stress Index 3
R1660 / R680
Apan, Held, Phinn and Markley (2003)
DWSI4
Disease Water Stress Index 4
R550 / R680
Apan, Held, Phinn and Markley (2003)
DWSI5
Disease Water Stress Index 5
(R800 + R550) / (R1660 + R680)
Apan, Held, Phinn and Markley (2003)
SR700_670
Simple Ratio 700/670
R700/R670 Part of TCARI index
RDVI
Renormalized Difference Vegetation Index
(R800 - R670)/(R800 + R670)^0.5
Rougean and Breon (1995)
PRI531
Normalized Difference 531/570 Photochemical Reflectance Index 531/570
(R531 - R570)/(R531 + R570)
Gamon et al. (1992)
EVI
Enhanced Vegetation Index
2.5 (R800 - R680) / (R800 + 6.0f R680 - 7.5f * R450 + 1.0f)
Huete et al. (1997)
ARVI
Atmospherically Resistant Vegetation Index
(R800 - (2.0f R680 - R450)) / (R800 + (2.0f R680 - R450))
Kaufman and Tanré (1996)
REIP1
Red-Edge Inflection Point 1
700 + 40 * {[(R670 + R780)/2 - R700] /(R740 - R700)}
Guyot and Baret, 1988
TVI
Triangular Vegetation Index
0.5 (120 (R750 - R550) - 200 * (R670 - R550))
Haboudaneet al. (2004)
GEMI
Global Environmental Monitoring Index
((2 (pow(R800) - pow(R680)) + 1.5 800 + 0.5 680) / (800 + 680 + 0.5) (1.0 - 0.25 (2.0f (pow(800) - pow(680)) + 1.5 800 + 0.5 680) / (800 + 680 + 0.5))) - ((680 - 0.125) / (1.0 - 680))
Pinty and Verstraete (1992)
GARI
Green Atmospherically Resistant Index
(R800 - (R550 - 1.7 (R450 - R680))) / (R800 + (R550 - 1.7 (R450 - R680)))
Gitelson et al. (1996)
DVI
Difference Vegetation Index
R800 - R680
Tucker et al. (1979)
GDVI
Green Difference Vegetation Index
R800 - R550
Sripada et al. (2006)
GNDVI
Green Normalized Difference Vegetation Index
(R800 - R550) / (R800 + R550)
Gitelson and Merzlyak (1998)
GRVI
Green Ratio Vegetation Index
R800 / R550
Sripada et al. (2006)
SR750_710
Simple Ratio 750/710 Zarco-Tejada & Miller 2001
R750/R710
Zarco-Tejada et al. (2001)
MSR705_445
Modified simple ratio 705/445
(R750 - R445)/(R705 - R445)
Sims and Gamon (2002)
WI
Water index
R900 - R970
Penuelas. et al. (1993)
Chl index
Chlorophyll index
R750/R550
Gitelson and Merzlyak (1994)
NDVI705
Normalized Difference 750/705 Chl NDI
(R750 - R705)/(R750 + R705)
Gitelson and Merzlyak (1994)
ChlDela
Chlorophyll content
(R540 - R590)/(R540 + R590)
Delaieux et al. (2014)
FRI2
Fluorescence ratio indices 2
R740/R800
Dobrowski et al. (2005)
NDVI1
Normalized Difference Vegetation Index1
(R800 - R670)/( R800 + R670)
Rouse et al. (1973)
FRI1
Fluorescence ratio index1
R690/R600
Dobrowski et al. (2005)
OSAVI
Optimized Soil Adjusted Vegetation Index
(1 + 0.16) * (R800 - R670)/(R800 + R670 + 0.16)
Rondeaux et al. (1996)
NDRE
Normalized Difference 790/720 Normalized difference red edge index
(R790 - R720)/(R790 + R720)
Barnes et al. (2000)
Car1Black
Carotenoid index from Blackburn 1998
R800/R470
Blackburn (1998)
SIPI
Structure intensive pigment index
(R800 - R450)/(R800 + R650)
Penuelas. et al. (1995)
AntGitelson
Anthocyanin (Gitelson)
(1/R550 - 1/R700) * R780
Gitelson et al.(2003,2006)
Car2Black
Carotenoid index 2 from Blackburn 1998
(R800 - R470)/(R800 + R470)
Blackburn (1998)
PRI586
Photochemical reflectance index from Panigada et al 2014
(R531 - R586)/(R531 + R586)
Panigada et al. (2014)
AntGamon
Anthocyanin from Gamon and Surfus 1999
R650/R550
Gamon and Surfus (1999)
CarChap
Carotenoid index (Chappelle)
R760/R500
Chappelle et al. (1992)
PRI512
Photochemical reflectance index from Hernandez-Clemente et al 2011
(R531- R512)/(R531 + R512)
Hern√°ndez-Clemente et al. (2011)
TCARI_OSAVI
Transformed Chlorophyll Absorption in Reflectance Index/Optimized Soil-Adjusted Vegetation Index: TCARI/OSAVI
TCARI/OSAVI
Haboudane et al. (2002)
IPVI
Infrared Percentage Vegetation Index
R800 / (R800 + R680)
Crippen et al. (1990)
NLI
Non-Linear Index
(pow(R800, 2) - R680) / (pow(R800, 2) + R680)
Goel and Qin (1994)
MNLI
Modified Non-Linear Index
((pow(R800, 2) - R680) * 1.5f) / (pow(R800, 2) + R680 + 0.5f)
Yang et al. (2008)
SAVI
Soil Adjusted Vegetation Index
(1.5f * (R800 - R680)) / (R800 + R680 + 0.5f)
Huete et al. (1988)
TDVI
Transformed Difference Vegetation Index
sqrt(0.5f + ((R800 - R680) / (R800 + R680)))
Bannari et al. (2002)
VARI
Visible Atmospherically Resistant Index
(R550 - R680) / (R550 + R680 - R450)
Gitelson et al. (2002)
RENDVI
Red Edge Normalized Difference Vegetation Index
(R750 - R705) / (R750 + R705)
Gitelson and Merzlyak (1994)
mRESR
Modified Red Edge Simple Ratio Index
(R750 - R445) / (R750 + R445)
Sims and Gamon (2002)
mRENDVI
Modified Red Edge Normalized Difference Vegetation Index
(R750 - R705) / (R750 + R705 - 2.0f * R445)
Sims and Gamon (2002)
VOG1
Vogelmann Red Edge Index 1
R740 / R720
Vogelmann et al. (1993)
VOG2
Vogelmann Red Edge Index 2
(R734 - R747) / (R715 + R726)
Vogelmann et al. (1993)
VOG3
Vogelmann Red Edge Index 3
(R734 - R747) / (R715 + R720)
Vogelmann et al. (1993)
MCARI
Modified Chlorophyll Absorption Reflectance Index
((R700 - R670) - 0.2f (R700 - R550)) (R700 / R670)
Daughtry et al. (2000)
MCARI1
Modified Chlorophyll Absorption Reflectance Index Improved 1
1.2f (2.5f (R790 - R670) - 1.3f * (R790 - R550))
Haboudane et al. (2004)
MCARI2
Modified Chlorophyll Absorption Reflectance Index Improved 2
(1.5f (2.5f (R800 - R670) - 1.3f (R800 - R550))) / sqrt(pow(2.0f R800 + 1.0f, 2) - 6.0f R800 - 5.0f sqrt(R670) - 0.5f)
Haboudane et al. (2004)
MTVI
Modified Triangular Vegetation Index
1.2f (1.2f (R800 - R550) - 2.5f * (R670 - R550))
Haboudane et al. (2004)
MTVI2
Modified Triangular Vegetation Index Improved
1.5f (1.2f (R800 - R550) - 2.5f (R670 - R550)) / sqrt(pow(2.0f R800 + 1.0f ,2) - (6.0f R800 - 5.0f sqrt(R670)) - 0.5f)
Haboudane et al. (2004)
GMI1
Gitelson and Merzlak Index 1
R750 / R550
Gitelson and Merzlak (1997)
GMI2
Gitelson and Merzlak Index 2
R750 / R700
Gitelson and Merzlak (1997)
Lic1
Lichtenthaler Index 1
(R790 - R680) / (R790 + R680)
Lichtenthaler et al. 1996
Lic2
Lichtenthaler Index 2
R440 / R690
Lichtenthaler et al. 1996
Lic3
Lichtenthaler Index 3
R440 / R740
Lichtenthaler et al. 1996
NDNI
Normalized Difference Nitrogen Index
(log(1.0f / R1510) - log(1.0f / R1680)) / (log(1.0f / R1510) + log(1.0f / R1680))
Fourty et al. (1996)
MSR
Modified Simple Ratio
((R800 / R680) - 1.0f) / (sqrt(R800 / R680) + 1)
Chen et al. (1996)
LAI
Leaf Area Index
3.618f ((2.5.0f (R800 - R680)) / (R800 + 6.0f R680 - 7.5.0f R450 + 1.0f)) - 0.118f
Boegh et al. (2002)
NRI1510
Nitrogen Related Index NRI1510
(R1510 - R660) / (R1510 + R660)
Herrmann et al. (2009)
NRI850
Nitrogen Related Index NRI850
(R850 - R660) / (R850 + R660)
Behrens et al. (2006)
NDLI
Normalized Difference Lignin Index
(log(1.0f / R1754) - log(1.0f / R1680)) / (log(1.0f / R1754) + log(1.0f / R1680))
Melillo et al. (1982)
CAI
Cellulose Absorption Index
(0.5 * (R2000 - R2200)) / R2100
Daughtry et al. (2001)
PSRI
Plant Senescence Reflectance Index
(R680 - R500) / R750
Merzlyak et al. (1999)
CRI1
Carotenoid Reflectance Index 1
1.0f / R510 - 1.0f / R550
Gitelson et al. (2002)
CRI2
Carotenoid Reflectance Index 2
1.0f / R510 - 1.0f / R700
Gitelson et al. (2002)
ARI1
Anthocyanin Reflectance Index 1
1.0f / R550 - 1.0f / R700
Gitelson et al. (2001)
ARI2
Anthocyanin Reflectance Index 2
R800 * ((1.0f / R550) - (1.0f / R700))
Gitelson et al. (2001)
SRPI
Simple Ration Pigment Index
R430 / R680
Penuelas et al. (1995)
NPQI
Normalized Phaeophytinization Index
(R415 - R435) / (R415 + R435)
Barnes et al. (1992)
NPCI
Normalized Pigment Chlorophyll Index
(R680 - R430) / (R680 + R430)
Penuelas et al. (1994)
WBI
Water Band Index
R900 / R970
Penuelas et al. (1995)
NDWI
Normalized Difference Water Index
(R857 - R1241) / ( R700 + R1241)
Gao et al. (1995)
MSI
Moisture Stress Index
R819 / R1599
Hunt and Rock (1989)
NDII
Normalized Difference Infrared Index
(R857 - R1241) / ( R700 + R1241)
Hardisky et al. (1983)
NMDI
Normalized Multiband Drought Index
(R819 - R1649) / (R819 + R1649)
Wang and Qu (2007)
HI
Healthy Index
((R534 - R698) / (R534 + R698)) - (R704 / 2.0f)
Mahlein et al. (2013)
CLSI
Cercospora Leaf Spot Index
((R698 - R570) / (R698 + R570)) - R734
Mahlein et al. (2013)
SBRI
Sugar Beet Rust Index
((R570 - R513) / (R570 + R513)) + (R704 / 2.0f)
Mahlein et al. (2013)
PMI
Powdery Mildew Index
((R520 - R584) / (R520 + R584)) + R724
Mahlein et al. (2013)
Crt1
Carter Index 1
R695 / R420
Carter (1994)
Crt2
Carter Index 2
R695 / R760
Carter (1996)
BIG2
Blue/Green Index
R450 / R550
Zarco-Tejada et al. (2005)
LSI
Leaf Structure Index
R1110 / R810
Maruthi Sridhar et al. (2007)
BRI
Browning Reflectance Index
((1.0f / R550) - (1.0f / R700)) / R800
Chivkunova et al. (2001)
G
Greenness Index
R554 / R677
Index
Label
Formula
Citation
DWSI1
Disease Water Stress Index 1
R800 / R1660
Apan, Held, Phinn and Markley (2003)
ND900_680
Normalized Difference 900/680
(R900 - R680)/(R900 + R680)
Rouse et al. (1973)
SR900_680
Simple ratio 900/680
traits::