Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Loading...
Several different sensors include geospatial information in the dataset metadata describing the location of the sensor at the time of capture.
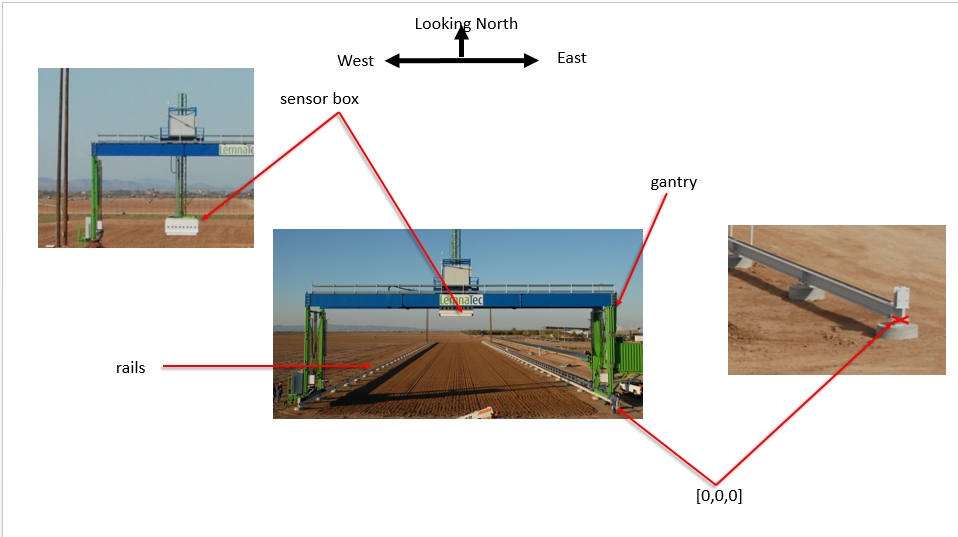
Coordinate reference systems The Scanalyzer system itself does not have a reliable GPS unit on the sensor box. There are 3 different coordinate systems that occur in the data:
Most common is EPSG:4326 (WGS84) USDA coordinates
Tractor planting & sensor data is in UTM Zone 12
Sensor position information is captured relative to the southeast corner of the Scanalyzer system in meters
EPSG:4326 coordinates for the four corners of the Scanalyzer system (bound by the rails above) are as follows:
NW: 33° 04.592' N, -111° 58.505' W
NE: 33° 04.591' N, -111° 58.487' W
SW: 33° 04.474' N, -111° 58.505' W
SE: 33° 04.470' N, -111° 58.485' W
In the trait database, this site is named the "MAC Field Scanner Field" and its bounding polygon is "POLYGON ((-111.9747967 33.0764953 358.682, -111.9747966 33.0745228 358.675, -111.9750963 33.074485715 358.62, -111.9750964 33.0764584 358.638, -111.9747967 33.0764953 358.682))"
Scanalyzer coordinates Finally, the Scanalyzer coordinate system is right-handed - the origin is in the SE corner, X increases going from south to north, and Y increases from east to the west.
In offset meter measurements from the southeast corner of the Scanalyzer system, the extent of possible motion for the sensor box is defined as:
NW: (207.3, 22.135, 5.5)
SE: (3.8, 0, 0)
Scanalyzer -> EPSG:4326 1. Calculate the UTM position of known SE corner point 2. Calculate the UTM position of the target point, using SE point as reference 3. Get EPSG:4326 position based on UTM
MAC coordinates Tractor planting data and tractor sensor data will use UTM Zone 12.
Scanalyzer -> MAC Given a Scanalyzer(x,y), the MAC(x,y) in UTM zone 12 is calculated using the linear transformation formula:
Assume Gx = -Gx', where Gx' is the Scanalyzer X coordinate.
MAC -> Scanalyzer
MAC -> EPSG:4326 USDA We do a linear shifting to convert MAC coordinates in to EPSG:4326 USDA
Sensors with geospatial metadata
stereoTop
flirIr
co2
cropCircle
PRI
scanner3dTop
NDVI
PS2
SWIR
VNIR
Available data All listed sensors
stereoTop
cropCircle
co2Sensor
flirIrCamera
ndviSensor
priSensor
SWIR
field scanner plots
There are 864 (54*16) plots in total and the plot layout is described in the plot plan table.
dimension
value
# rows
32
# rows / plot
2
# plots (2 rows ea)
864
# ranges
54
# columns
16
row width (m)
0.762
plot length (m)
4
row length (m)
3.5
alley length (m)
0.5
The boundary of each plot changes slightly each planting season. The scanalyzer coordinates of each row and each range of the two planting seasons is available in the field book. The scanalyzer coordinates of each plot are transformed into the (EPSG:4326) USDA coordinates using the equations above. After that, a polygon of each plot can be generated using ST_GeomFromText funtion and inserted into the BETYdb through SQL statements.
An Rcode is available for generating SQL statements based on the scanalyzer coordinates of each plot, which takes range.csv and row.csv as standard inputs.
The range.csv should be in the following format:
range
x_south
x_north
1
...
...
2
...
...
3
...
...
...
...
...
And the row.csv should look like:
row
y_west
y_east
1
...
...
2
...
...
3
...
...
...
...
...
The output will be something look like:
Genomic data includes whole-genome resequencing data from the HudsonAlpha Institute for Biotechnology, Alabama for 384 samples for accessions from the sorghum Bioenergy Association Panel (BAP) and genotyping-by-sequencing (GBS) data from Kansas State University for 768 samples from a population of sorghum recombinant inbred lines (RIL).
These data are available to Beta Users and require permission to access. The form to sign up for our beta user program is at http://terraref.org/beta. Once you have signed up for our beta user program you can access genomics data in one of the following locations:
Download via Globus.
The NDS Analysis Workbench, which provides container-based computing environments including Jupyter, Rstudio, and Python IDE.
The CyVerse Data Store for download or use within the CyVerse computing environment.
The CoGe computing environment.
See Data use policy before continuing.
The data is structured on both the TERRA-REF strorage (accessible via Globus and Workbench) and CyVerse Data Store infrastructures as follows:
Raw data are in bzip2 FASTQ format, one per read pair (*_R1.fastq.bz2 and *_R2.fastq.bz2). 384 samples are available. For a list of the lines sequenced, see the sample table.
Data derived from analysis of the raw resequencing data at the Danforth Center (version1) are available as gzipped, genotyped variant call format (gVCF) files and the final combined hapmap file.
Raw data are in gzip FASTQ format. 768 samples are available. For a list of lines sequenced, see the sample table.
Combined genotype calls are available in VCF format.
Meteorological data will use Climate Forecasting 'standard names' and 'canonical units' conventions. CF is widely used in climate, meteorology, and earth sciences.
Here are some examples (note that we can change from canonical units to match the appropriate scale, e.g. "C" instead of "K"; time can use any base time and time step (e.g. hours since 2015-01-01 00:00:00 UTC, etc. But the time zone has to be UTC, where 12:00:00 is approx (+/- 15 min). solar noon at Greenwich.
CF standard-name
units
time
days since 1700-01-01 00:00:00 UTC
air_temperature
K
air_pressure
Pa
mole_fraction_of_carbon_dioxide_in_air
mol/mol
moisture_content_of_soil_layer
kg m-2
soil_temperature
K
relative_humidity
%
specific_humidity
1
water_vapor_saturation_deficit
Pa
surface_downwelling_longwave_flux_in_air
W m-2
surface_downwelling_shortwave_flux_in_air
W m-2
surface_downwelling_photosynthetic_photon_flux_in_air
mol m-2 s-1
precipitation_flux
kg m-2 s-1
irrigation_flux
kg m-2 s-1
irrigation_transport
kg s-1
wind_speed
m/s
eastward_wind
m/s
northward_wind
m/s
standard_name is CF-convention standard names (except irrigation)
units can be converted by udunits, so these can vary (e.g. the time denominator may change with time frequency of inputs)
Before the Running
The pipepline is developed in Python, so a Python Interpreter is a must. Other than the basic Python standard librarys, the following third-party libraries are required:
netCDF4 for Python
numpy
Other than official CPython interpreter, Pypy is also welcomed, but please make sure that these third-party modules are correctly installed for the target interpreter. The pipeline can only works in Python 2.X versions (2.7 recommended) since numpy does not support Python 3.X versions.
Cloning from the Git:
The extractor for this pipeline is developed and maintained by Max in branch "EnvironmentalLogger-extractor" under the same repository.
Get the Environmental Logger Pipeline to Work
To trigger the pipeline, use the following command:
python ${environmental_logger_source_path}/environmental_logger_json2netcdf.py ${input_JSON_file} ${output_netCDF_file}
Where:
${environmental_logger_source_path} is where the three environmental_logger files are located
${input_JSON_file} is where the input JSON files are located
${output_netCDF_file} is where the users want the pipeline to export the product (netCDF file)
Please note that the parameter for the output file can be a path to either a directory or a file, and it is not necessarily to be existed. If the output is a path to a folder, the final product will be in this folder as a netCDF file that has the same name as the imported JSON file but with a different filename extension (.nc for standard netCDF file); if this path does not exist, environmental_logger pipeline will automatically make one.
The calculation in the Environmental Logger is mainly finished by the module environmental_logger_calculation.py under the support of numpy.
Environment conditions data is collected using the Vaisala CO2, Thies Clima weather sensors as well as lightning, irrigation, and weather data collected at the Maricopa site.
Data formats follow the Climate and Forecast (CF) conventions for variable names and units. Environmental data are stored in the Geostreams database.
WeatherStation coordinates are 33.074457 N, 111.975163 W
EnvironmentLogger is on top of the gantry system and is moveable.
Irrigation is managed at the field level. There are four regions that can be irrigated at different rates.
Level 1 meteorological data is aggregated to from 1 Hz raw data to 5 minute averages or sums.
On Globus or Workbench you can find these data provided in both hourly and daily files. These files contain data at the original temporal resolution of 1/s. In addition, they contain the high resolution spectral radiometer data.
sites/ua-mac/Level_1/envlog_netcdf
hourly files: YYYY-MM-DD_HH-MM-SS_environmentallogger.nc
daily files: envlog_netcdf_L1_ua-mac_YYYY-MM-DD.nc
Data can be accessed using the geostreams API or the PEcAn meteorological workflow. These are illustrated in the sensor data tutorials.
Here is the json representation of a single five-minute observation:
Data can be accessed using the geostreams API or the PEcAn meteorological workflow.
These are illustrated in the sensor data tutorials.
Here is the json representation of a single five-minute observation from Geostreams:
CF standard-name
units
bety
isimip
cruncep
narr
ameriflux
air_temperature
K
airT
tasAdjust
tair
air
TA (C)
air_temperature_max
K
tasmaxAdjust
NA
tmax
air_temperature_min
K
tasminAdjust
NA
tmin
air_pressure
Pa
air_pressure
PRESS (KPa)
mole_fraction_of_carbon_dioxide_in_air
mol/mol
CO2
moisture_content_of_soil_layer
kg m-2
soil_temperature
K
soilT
TS1 (NOT DONE)
relative_humidity
%
relative_humidity
rhurs
NA
rhum
RH
specific_humidity
1
specific_humidity
NA
qair
shum
CALC(RH)
water_vapor_saturation_deficit
Pa
VPD
VPD (NOT DONE)
surface_downwelling_longwave_flux_in_air
W m-2
same
rldsAdjust
lwdown
dlwrf
Rgl
surface_downwelling_shortwave_flux_in_air
W m-2
solar_radiation
rsdsAdjust
swdown
dswrf
Rg
surface_downwelling_photosynthetic_photon_flux_in_air
mol m-2 s-1
PAR
PAR (NOT DONE)
precipitation_flux
kg m-2 s-1
cccc
prAdjust
rain
acpc
PREC (mm/s)
degrees
wind_direction
WD
wind_speed
m/s
Wspd
WS
eastward_wind
m/s
eastward_wind
CALC(WS+WD)
northward_wind
m/s
northward_wind
CALC(WS+WD)
Data is available via Globus or Workbench:
/ua-mac/raw_data/co2sensor
/ua-mac/raw_data/EnvironmentLogger
/ua-mac/raw_data/irrigation
/ua-mac/raw_data/lightning
/ua-mac/raw_data/weather
Description: EnvironmentalLogger raw files are converted to netCDF.
Known issue: the irrigation data stream does not currently handle variable irrigation rates within the field. Specifically, we have not yet accounted for the Summer 2017 drought experiments. See terraref/reference-data#196 for more information.
When the full field is irrigated (as is typical), the irrigated area is 5466.1 m2 (=215.2 m x 25.4 m)
In 2017:
Full field irrigated area from the start of the season to August 1 (103 dap) is 5466.1 m2 (=215.2 m x 25.4 m).
Well-watered treatment zones from August 1 to 15 (103 to 116 dap): 2513.5 m2 (=215.2 m x 11.68 m) in total, combined areas of non-contiguous blocks
Well-watered treatment zones from August 15 - 30 (116 to 131 dap): 3169.9 m2 (=215.2 m x 14.73 m), again in total as the combined areas of non-contiguous blocks
3D point cloud data is collected using the Fraunhofer 3D laser scanner. Custom software installed at Maricopa converts .png output to the .ply point clouds. The .ply point clouds are converted to georeferenced .las files using the 3D point cloud extractor
Level 1 data products are provided in both .las and .ply formats.
For each scan, there are two .ply files representing two lasers, one on the left and the other on the right. These are combined in the .las files.
For details about using this data via Clowder or Globus, please see Data Access section.
Data is available via Clowder, Globus, and Workbench.
Clowder: Laser Scanner 3D LAS
Globus or Workbench File System:
LAS /sites/ua_mac/raw_data/laser3D_las
PLY /sites/ua_mac/raw_data/laser3D
Raw sensor output (PLY) is converted to LAS format using the ply2las extractor
The position of the lasers is affected by temperature. We plan to add a correction for temperature that will adjust for this effect. See terraref/reference-data#161
Phenotype data is derived from images generated by the indoor LemnaTec Scanalyzer 3D platform at the Donald Danforth Plant Science Center using PlantCV. PlantCV is an image analysis package for plant phenotyping. PlantCV is composed of modular functions in order to be applicable to a variety of plant types and imaging systems. PlantCV contains base functions that are required to examine images from an excitation imaging fluorometer (PSII), visible spectrum camera (VIS), and near-infrared camera (NIR). PlantCV is a fully open source project: https://github.com/danforthcenter/plantcv. For more information, see:
Project website: http://plantcv.danforthcenter.org
Full documentation: http://plantcv.readthedocs.io/en/latest
Publications:
To learn more about PlantCV, you can find examples in the terraref/tutorials repository, which is accessible on GitHub and in the TERRA REF workbench under tutorials/plantcv
an ipython notebook demonstration of PlantCV plantcv/plantcv_jupyter_demo.ipynb.
For the TERRA-REF project, a PlantCV Clowder extractor was developed to analyze data from the Bellwether Foundation Phenotyping Facility at the Donald Danforth Plant Science Center. Resulting phenotype data is stored in BETYdb.
Description: Processes VIS/NIR images captured at several angles to generate trait metadata. The trait metadata is associated with the source images in Clowder, and uploaded to the configured BETYdb instance.
Output CSV: /sites/danforth/Level_1/<experiment name>
Input
Evaluation is triggered whenever a file is added to a dataset
Following images must be found
2x NIR side-view = NIR_SV_0, NIR_SV_90
1x NIR top-view = NIR_TV
2x VIS side-view = VIS_SV_0, VIS_SV_90
1x VIS top-view = VIS_TV
Per-image metadata in Clowder is required for BETYdb submission; this is how barcode/genotype/treatment/timestamp are determined.
Output
Each image will have new metadata appended in Clowder including measures like height, area, perimeter, and longest_axis
Average traits for the dataset (3 VIS or 3 NIR images) are inserted into a CSV file and added to the Clowder dataset
If configured, the CSV will also be sent to BETYdb
For details about accessing BETYdb, please see Data Access section and a tutorial on accessing phenotypes from the trait database on the TERRA REF Workbench in traits/04-danforth-indoor-phenotyping-facility.Rmd.
Globus and Workbench:
/sites/danforth/raw_data/<experiment name>
Fluorescence intensity data is collected using the PSII camera.
Fluorescence intensity data is available via Clowder and Globus:
Clowder: __ps2Top collection
Globus path: /sites/ua-mac/raw_data/ps2top
Sensor information: LemnaTec PSII
For details about using this data via Clowder or Globus, please see Data Access section.
Description: Raw image output is converted to a raster format (netCDF\/GeoTIFF)
Output: /sites/ua_mac/Level_1/ps2top
There are 102 bin files. The first (index 0) is an image taken right before the LED are switched on (dark reference). Frame 1 to 100 are the 100 images taken, with the LEDs on. In binary file 102 (index 101) is a list with the timestamps of each frame of the 100 frames.
Right now the LED on timespan is 1s thus the first 50 frames are taken with LEDs on the latter 50 frames with LED off..

The following table lists available TERRA-REF data products. The table will be updated as new datasets are released. Links are provided to pages with detailed information about each data product including sensor descriptions, algorithm (extractor) information, protocols, and data access instructions.
Data product
Description
3D point cloud data (LAS) of the field constructed from the Fraunhofer 3D scanner output (PLY).
Fluorescence intensity imaging is collected using the PSII LemnaTec camera. Raw camera output is converted to (netCDF/GeoTIFF)
Hyperspectral imaging data from the SWIR and VNIR Headwall Inspector sensors are converted to netCDF output using the hyperspectral extractor.
Infrared heat imaging data is collected using FLIR sensor. Raw output is converted to GeoTIFF using the FLIR extractor.
Multispectral data is collected using the PRI and NDVI Skye sensors. Raw output is converted to timeseries data using the multispectral extractor.
Stereo imaging data is collected using the Prosilica cameras. Full-color images are reconstructed in GeoTIFF format using the de-mosaic extractor. A full-field mosaic is generated using the full-field mosaic extractor.
Spectral reflectance data
Spectral reflectance is measured using a Crop Circle active crop canopy sensor
Environment conditions are collected through the CO2 sensor and Thies Clima. Raw output is converted to netCFG using the environmental-logger extractor.
postGIS/netCDF
Phenotype data is derived from sensor output using the PlantCV extractor and imported into BETYdb.
FASTQ and VCF files available via Globus
UAV and Phenotractor
Plot level data available in BETYdb
Hyperspectral imaging data is collected using the Headwall VNIR and SWIR sensors. In the Nov 2017 Beta Release only VNIR data is provided because we do not have the measurements of downwelling spectral radiation required by the pipeline.
Please see the for more information about how the data are generated and known issues.
See
Hyperspectral data is available via Clowder, , the , and our :
Clowder:
SWIR Collection: Level 1 data not available
Globus and Workbench:
VNIR: /sites/ua-mac/Level_1/vnir_netcdf
SWIR: Level 1 data not available
Sensor information:
Level 2 data are spectral indices computed at the same resolution as Level 1. These can be found in the same Level 1 directories as their parents, but the files are appended *_ind.nc.
To get a list of hyperspectral indices currently generated:
Raw data is available in the filesystem, accessible via Workbench and Globus in the following directories:
VNIR: /sites/ua-mac/raw_data/VNIR
SWIR: /sites/ua-mac/raw_data/SWIR
These files are uncalibrated; see the hyperspectral pipeline repository for information on how these can be processed.
Infrared heat imaging data is collected collected using the FLIR SC615 thermal sensor. These data are provided as geotiff image raster files as well as plot level means.
Algorithms are in the repository; see the readme for details.
Sensor information:
ua-mac/Level_1/ir_geotiff
To be created
Plot level summaries are named in the trait database. In the future this name will be used for the Level 1 data as well. This name from the Climate Forecast (CF) conventions, and is used instead of 'canopy_temperature' for two reasons: First, because we do not (currently) filter soil in this pipeline. Second, because the CF definition of surface_temperature distinguishes the surface from the medium: "The surface temperature is the temperature at the interface, not the bulk temperature of the medium above or below."
Thermal imaging data is available via Clowder and Globus:
/ua-mac/raw_data/flirIrCamera
Data are unavailable for Season 4 (summer 2017 sorghum) and season 5 (winter 2017-2018 wheat).
For details about using this data via Clowder or Globus, please see section.
For details about using this data via Clowder or Globus, please see section.
Work to recover these data is ongoing; see
Problem description